DIVISION OF GENOMIC RESOURCES

Head of Division: Dr. Rakesh Singh
Contact Numbers: 011-25802884; Ext. 791 Fax:011-25843697; Mob +91 9818934006
Address: ICAR-National Bureau of Plant Genetic Resources, Pusa Campus, New Delhi – 110012, INDIA
E-mail: akesh.singh2-icar@nic.in, singhnbpgr@yahoo.com
Google Map
Mandate
The National Research Centre on DNA Fingerprinting was established by the Indian Council of Agricultural Research in December 1995 at ICAR-National Bureau of Plant Genetic Resources (NBPGR), which was renamed as the Division of Genomic Resources w.e.f. 20th February 2013. The division has been involved in activities, generation, utilization, and conservation of plant genomic resources, with the following mandate:
- To manage and promote sustainable use of plant genetic and genomic resources of agri-horticultural crops and carry out related research
- Molecular profiling of varieties of agri-horticultural crops and GM detection technology research
Salient Achievements
- Cereals, Pseudocereals and Millets
2. The genome-wide SSRs were developed and validated for Amaranthus hypochondriacus. New genomic resources were generated for amaranth characterization and future breeding programs in India. ( https://doi.org/10.3390/agriculture13020431)
- Oilseeds and Pulses
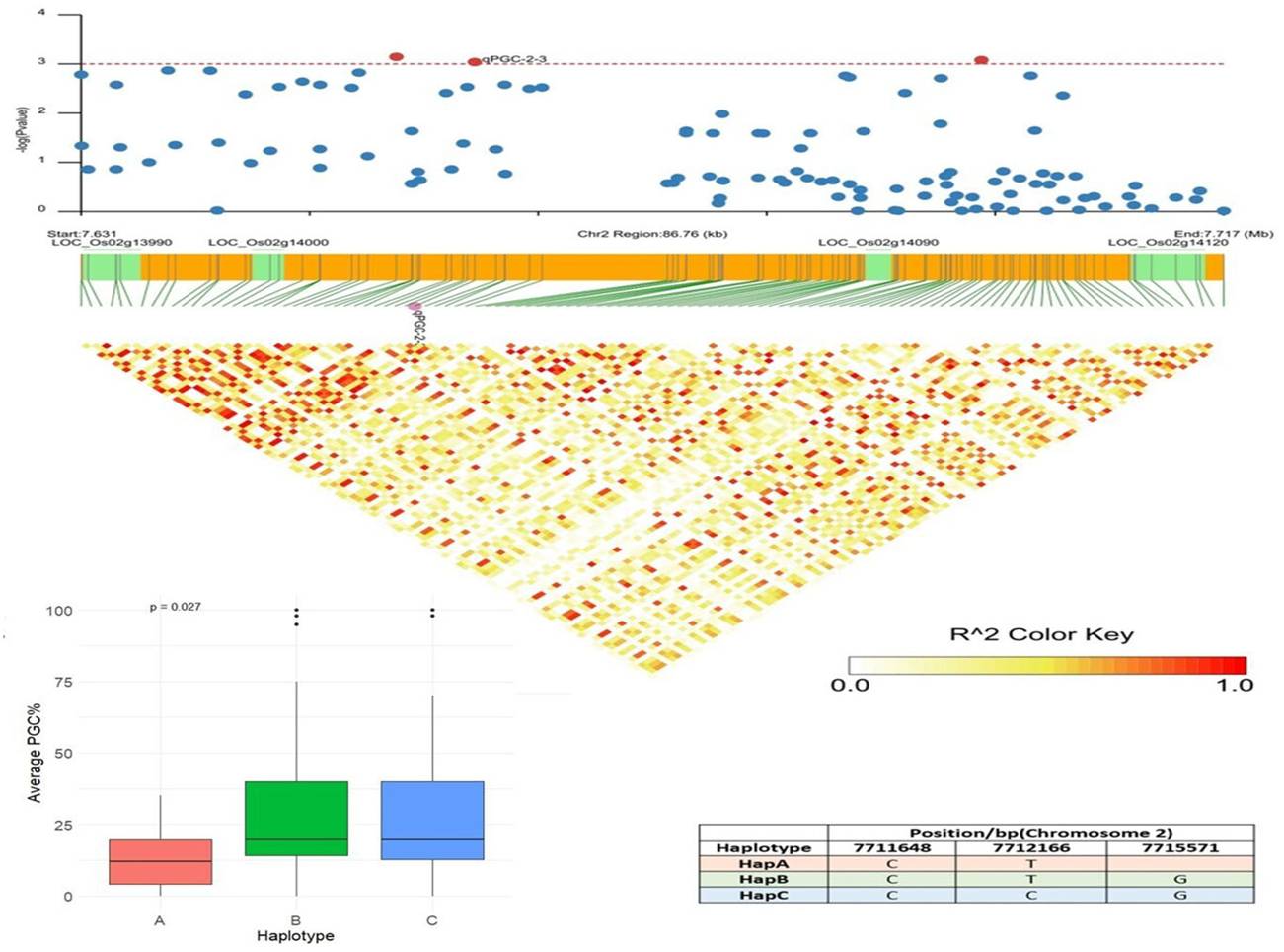
1. Genomic resources in the form of 68,925 SNPs were developed using genotyping by sequencing (GBS) of 131 linseed germplasm accessions conserved in the Indian National Genebank (INGB).
2. Quantitative trait nucleotides (QTNs) were identified in linseed using Genome-Wide Association Study (GWAS) for flowering time (53 QTNs), days to maturity (30 QTNs), plant height (27 QTNs), and thousand seed weight (30 QTNs) in linseed.
(A). Manhattan plots and quantile-quantile plots for days to 5%(DF5), 50% (DF50), and 95% flowering (DF95), days to maturity (DM), and plant height (PH) using five ML-GWAS methods. The dotted lines in Manhattan plots show a threshold at LOD score of ≥3.0. The pink dots depict significant QTNs identified by ≥2 methods. (B) Chromosomal positions of stable QTNs for flowering time (DF5, DF50, and DF95) (blue dots), days to maturity (DM) (red dots), and plant height (PH) (green dots) traits in linseed.
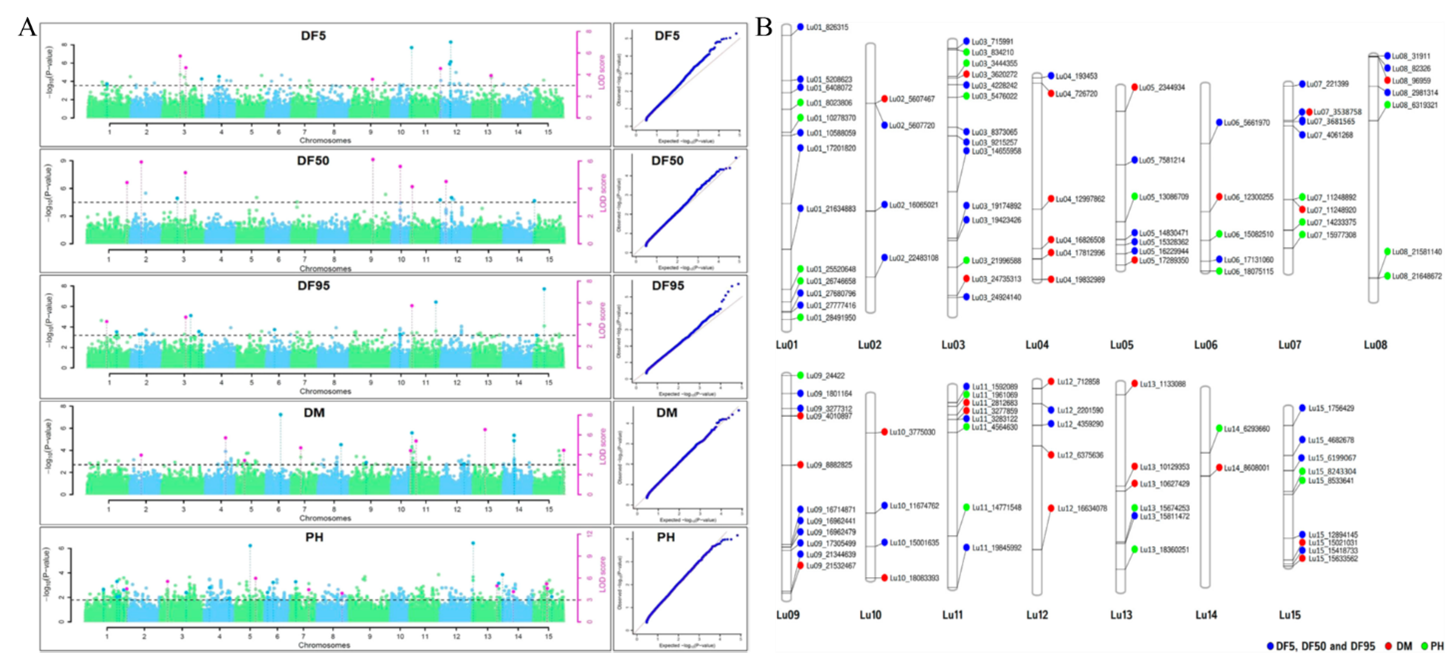
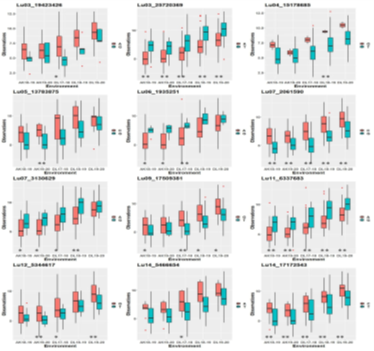
3. For Thousand Seed Weight (TSW) in linseed a total of 30 stable QTNs were identified accounting for up to 38.65% trait variation. Alleles with a positive effect on trait were analyzed for 12 strong QTNs with r2 ≥ 10.00%, which showed a significant association of specific alleles with higher trait values in three or more environments.
- Horticultural Crops
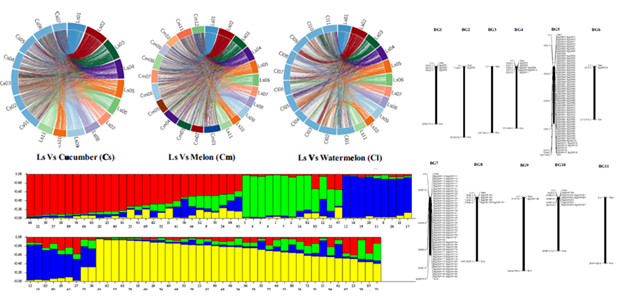
- Development of genome-wide SSR markers in bottle gourd, study of synteny of mapped bottle gourd SSRs markers with other cucurbits, viz cucumber, watermelon and melon and population structure analyses in National GeneBank bottle gourd germplasm (https://doi.org/10.1007/s13353-022-00684-1)
2. Variability in sponge gourd genotypes for biochemical parameters and identification of genotypes with improved oil and nutrient quality (https://doi.org/10.3389/fnut.2023.1158424)
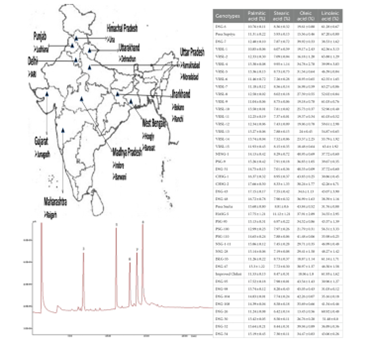
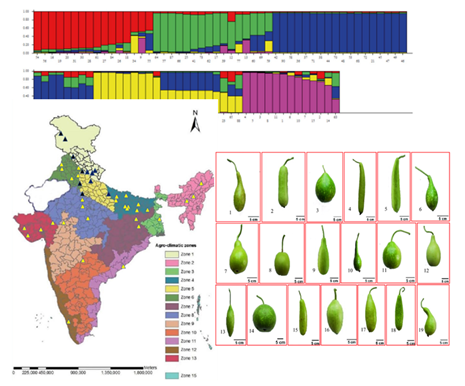
3. Assessment of genetic diversity in bottle gourd from different agro-climatic zones of India using agro-morphological traits and SSR markers. (https://doi.org/10.1007/s11033-022-07446-6)
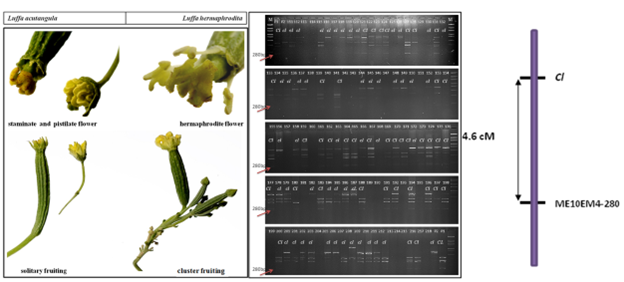
4. Elucidation of inheritance pattern of fruiting behaviour in Luffa, designation of the gene symbol clfor cluster fruit bearing in Luffa and linkage analysis of SRAP marker ME10 EM4-280 linked to the fruiting trait (Cl locus) (https://doi.org/10.1007/s00438-023-02034-0)
5. Genetic Diversity and Population Structure Studies: 3115 safflower accessions from the Indian National Gene Bank, including Indian cultivars were assessed for genetic diversity and population structure using 18 polymorphic SSR markers ( https://doi.org/10.3390/agriculture13040836). Similar studies were carried out in Coixlacryma-jobi and barley.
6. For the first time, cryobionomics of Banana (Musa sp. AAA cv Borjahaji), a non-model species, was investigated through comprehensive transcript profiling during four stages of cryopreservation (https://doi.org/10.3390/plants12051165).
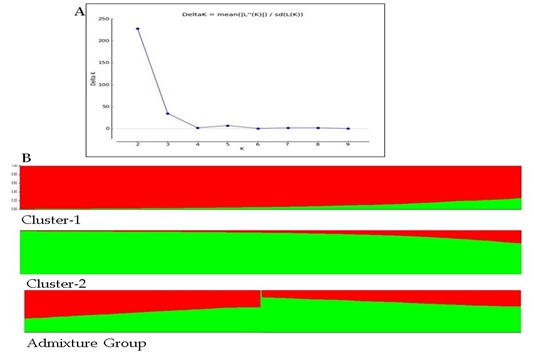
7. Genetic diversity in Morinda spp. for its enhanced collection, conservation and utilization (https://doi.org/10.1007/s10722-021-01321-2)
B. Molecular Systematics
1. DNA Barcode for Species Identification: DNA barcoding was generated for wild and cultivated species of Luffa, vegetable Amaranthus species complex, Trichosanthes cucumerina complex, Cucumis melo, Brinjal, Allium and wild Musa.
2. Molecular cytogenetics and Flow Cytometry:
- Molecular cytogenetics is utilized in species identification in Vegetable Amaranthus and Cucumber.
- In wild Musa species flow cytometry technique was utilized for their genome size estimation and ploidy determination, which was complemented with cytogenetic studies (https://doi.org/10.3390/plants12203605), which helped in establishing the identity of samples conserved at the tissue culture of ICAR-NBPGR.
- Protocol for Fluorescence in situ hybridization (FISH) was standardized for Musa
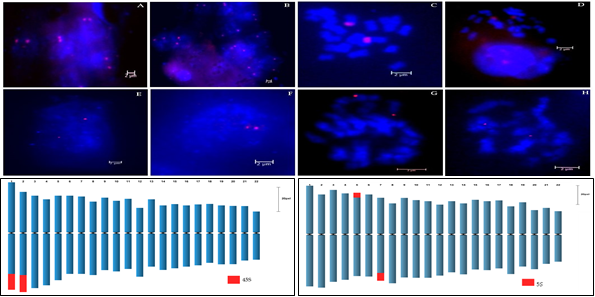
- Chromosome identification and karyotype analysis of Vigna umbellata was carried out using Fluorescence in situ hybridization.
C. Whole Genome Sequencing
- The de novoassembled draft whole genome sequence of small cardamom, (Eletaria cardamomum. Maton ) has been reported (https://doi.org/10.3389/fpls.2023.1161499)
- Rice Core: SNP markers-based North-East Core (https://doi.org/10.1371/journal.pone.0113094)and East Coast Core (https://doi.org/10.3389/fgene.2021.726152)
- Niger Core: Agronomic traits-based core set.
- Safflower Core: SNP markers-based core set.
- Cowpea Core: Agronomic traits-based core set.
- Horse gram Core: Agronomic traits-based core set.
- Black gram Core: Agronomic traits-based core set.
- Green gram Core: Agronomic traits-based core set.
- Sesame Core: SNP markers-based core set.
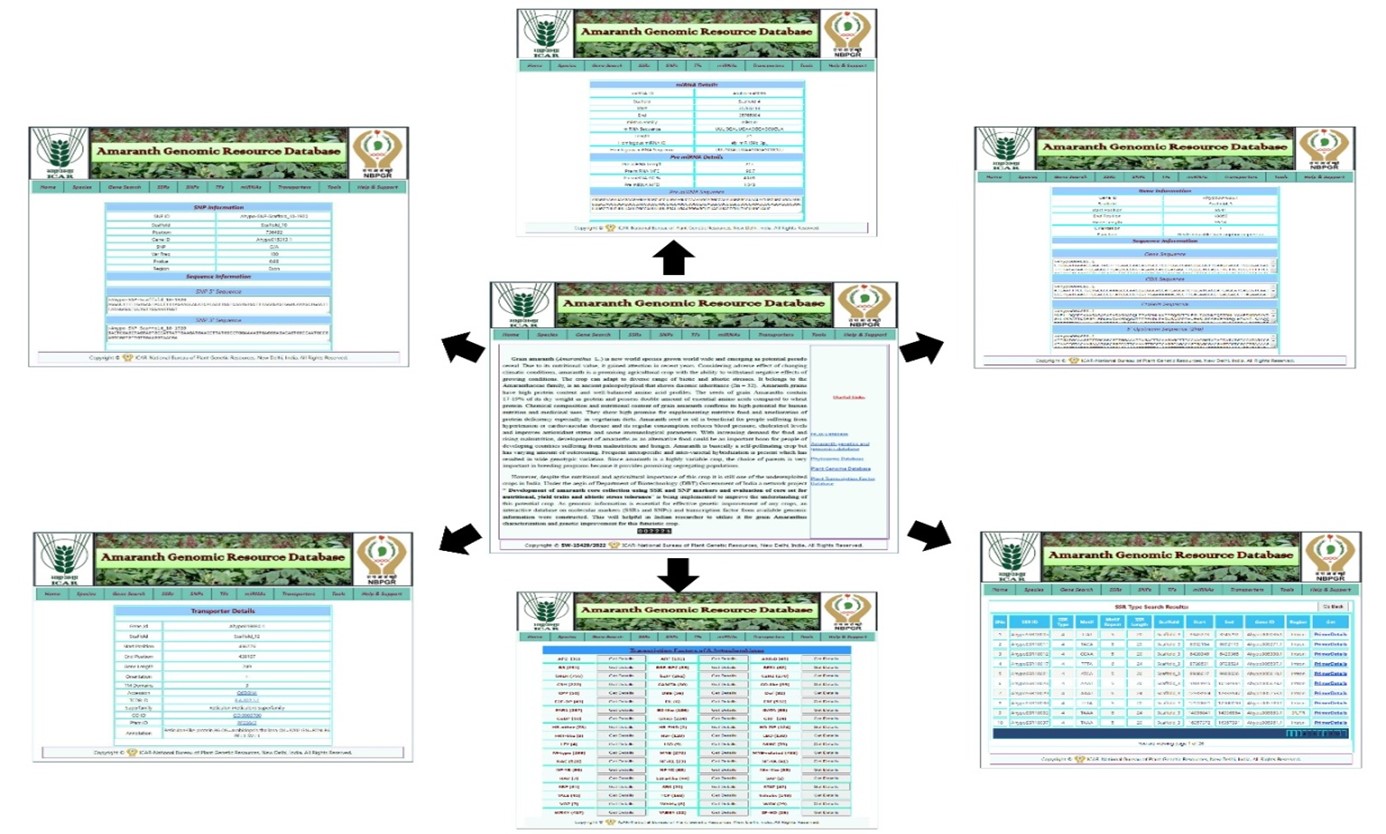
D. Database Development
- Amaranth Genomic Resource Database (AGRDB) (http://www.nbpgr.ernet.in:8080/AmaranthGRD/).
- Andrographis paniculata Genomic Resource Database (https://doi.org/10.3390/ijms24119212)
- Tinospora cordifolia genomic Resource Database (https://doi.org/10.3390/genes13081433)
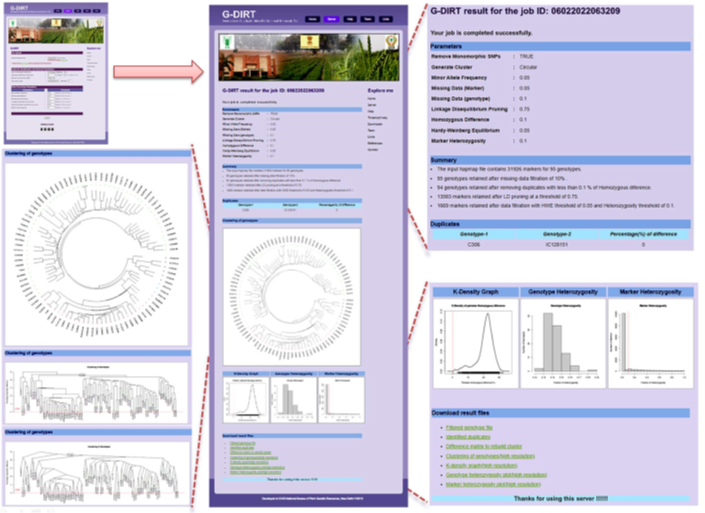
- A web server “Germplasm Duplicate Identification and Removal Tool” (G-DIRT) has been developed which allows the identification of duplicate germplasm based on SNP genotyping data. (http://webtools.nbpgr.ernet.in/gdirt/)
| Table: DNA fingerprinting status of varietal sample during last five years | |||||||
| Crop | Scientific Name | 2019-20 | 2020-21 | 2021-22 | 2022-23 | 2023-24 | Total |
| Amaranth | Amaranthus sp. | 3 | 3 | ||||
| Barley | Hordeum vulgare | 1 | 1 | ||||
| Bitter gourd | Momordica charantia | 1 | 1 | ||||
| Bottle gourd | Lagenaria siceraria | 1 | 1 | ||||
| Brinjal | Solanum melongena | 9 | 4 | 1 | 1 | 15 | |
| Buckwheat | Fagopyrum esculentum | 1 | 1 | 2 | |||
| Chenopodium | Chenopodium | 1 | 1 | ||||
| Chickpea | Cicer arietinum | 2 | 1 | 3 | |||
| Chilli | Capsicum annum | 2 | 1 | 1 | 4 | ||
| Cotton | Gossypium sp. | 21 | 59 | 5 | 12 | 9 | 106 |
| Cowpea | Vigna unguiculata | 2 | 3 | 5 | |||
| Dolichos bean | Dolichos | 1 | 1 | ||||
| Faba bean | Vicia faba | 1 | 1 | ||||
| Fennel | Foeniculum vulgare | 1 | 1 | ||||
| Fenugreek | Trigonella foenum-graecum | 1 | 1 | ||||
| Finger millet | Eleusine coracana | 1 | 1 | ||||
| Foxtail millet | Setaria italica | 1 | 1 | 2 | |||
| Frenchbean | Phaseolus vulgaris | 4 | 4 | ||||
| Horsegram | Macrotyloma uniflorum | 4 | 4 | ||||
| Hot pepper | Capsicum frutescens | 6 | 6 | ||||
| Lentil | Lens culinaris | 2 | 2 | ||||
| Linseed | Linum usitatissimum | 1 | 1 | 2 | |||
| Little millet | Panicum sumatrense | 2 | 2 | ||||
| Maize | Zea mays L. | 10 | 3 | 6 | 19 | ||
| Mung bean | Vigna radiata | 4 | 5 | 7 | 3 | 19 | |
| Muskmelon | Cucumis melo | 1 | 1 | ||||
| Mustard | Brassica sp. | 5 | 7 | 3 | 4 | 19 | |
| Oats | Avena sativa | 5 | 4 | 1 | 1 | 11 | |
| Okra | Abelmoschus esculentus | 4 | 4 | ||||
| Paddy | Oryza sativa | 26 | 23 | 1 | 27 | 2 | 79 |
| Pea | Pisum sativum | 1 | 3 | 1 | 5 | ||
| Pearl millet | Pennisetum glaucum | 4 | 4 | 4 | 4 | 16 | |
| Pigeon pea | Cajanus cajan | 4 | 13 | 17 | |||
| Raya | Eruca sp. | 2 | 1 | 3 | |||
| Ridge gourd | Luffa acutangula | 1 | 1 | ||||
| Sesame | Sesamum indicum | 6 | 1 | 1 | 8 | ||
| Sorghum | Sorghum bicolor | 1 | 5 | 6 | |||
| Soybean | Glycine max | 4 | 2 | 2 | 3 | 3 | 14 |
| Sponge gourd | Luffa aegyptiaca | 3 | 1 | 4 | |||
| Sunflower | Helianthus annuus | 1 | 1 | ||||
| Taramira | Eruca sativa | 1 | 1 | ||||
| Tomato | Solanum lycopersicum | 6 | 3 | 9 | |||
| Toria | Raphanus sativus | 5 | 5 | ||||
| Urd bean | Vigna mungo | 1 | 3 | 11 | 7 | 22 | |
| Walnut | Juglans regia | 8 | 2 | 2 | 12 | ||
| Watermelon | Citrullus lanatus | 1 | 1 | ||||
| Wheat | Triticum aestivum | 5 | 1 | 1 | 14 | 2 | 23 |
| Total | 117 | 153 | 51 | 109 | 39 | 469 | |
| GMO Testing Services |
| As a Referral Laboratory to detect presence or absence of Genetically Modified Organisms (GMO) notified under sub-section (1) of Section 4 of the Seeds Act, 1966 in a Gazette of India Notification (Department of Agriculture and Farmers Welfare, DAFW) in 2017, the GM detection research facility is rendering GMO testing services. Resource generated (2018-2023): Approx ₹15.00 lakhs |
| ICAR-NBPGR facilitates import of transgenics for research purpose (including their molecular testing) as per Government of India notification no. GSR 1067 (E) dated 5 Dec 1989, and Plant Quarantine (Regulation of Import into India) Order 2003 after technical clearance for import by RCGM. So far, >250 imported GM consignments have been tested. |
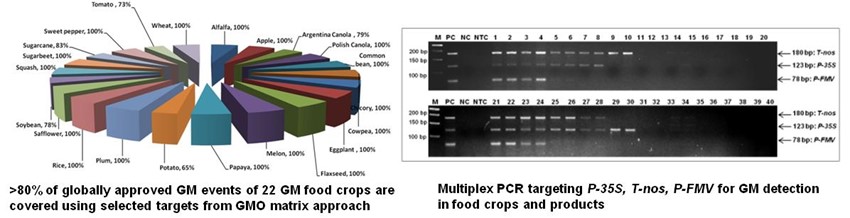
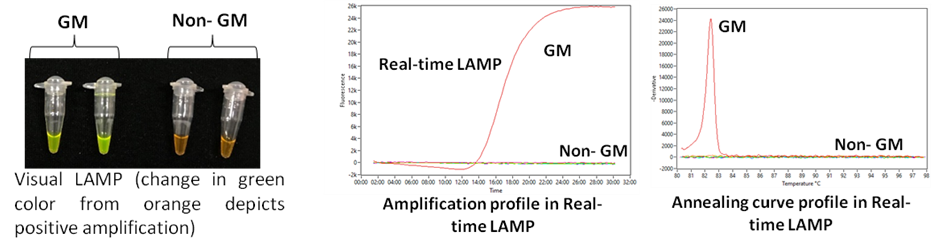
2. Loop-mediated Isothermal Amplification (LAMP) technology for rapid/on-site GM detection: Visual/Real-time LAMP assays developed for >25 targets (https://doi.org/10.1016/j.foodcont.2019.106759; https://doi.org/10.1007/s00217-023-04294-x)
Patents/ Copyrights /Commercialization of Technologies
Patents
- Patent No. 245749: Process enabling simultaneous detection of two transgenes namely 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS or CP4EPSPS) gene and cauliflowerInventors: Randhawa GJ, Firke PK and Karihaloo JL Commercialization: Available on Non-exclusive basis
- Patent No. 254341: Process enabling simultaneous detection of two transgenes namely human serum albumin (HAS) and bar genes using a multiplex polymerase chain reaction Inventors: Randhawa GJ, Firke PK and Karihaloo JL Commercialization: Available on Non-exclusive basis
- Patent No 258165: Diagnostic kit based on polymerase chain reaction for detection of cry1Ac geneInventors: Randhawa GJ and Firke PKCommercialization: Available on Non-exclusive basis
Copyrights
- Copyright Reg.No. 5760/06-CD (SW): Development of Crop DNA Fingerprinting Database Software Package Author: Madhu Bala Commercialization: Available on request for research use; for commercial use on non-exclusive basis
- Copyright Reg. No. SW-15429/2022: AMARANTH GENOMIC RESOURCE DATABASE Authors: Rakesh Singh, Akshay Singh, Ajay Kumar Mahato, S Rajkumar, A K Singh, Rakesh Bhardwaj, S K Kaushik, Sandeep Kumar and Veena Gupta Commercialization: Freely Available for non-commercial purposes.
- Copyright Reg. No. SW-15428/2022: TINOTRANSCRIPTDB (TINOSPORA CORDIFOLIA TRANSCRIPTS & SSR DATABASE) Authors: Rakesh Singh, Akshay Singh, Ajay Kumar Mahato, Rajesh Kumar, Amit K. Singh, Sundeep Kumar, Soma S. Marla, Ashok Kumar and Nagendra K. SinghCommercialization: Freely Available for non-commercial purposes.
- Copyright Reg. No. SW-15900/2022: APTRANSDB: ANDROGRAPHIS TRANSCRIPTS & SSR DATABASEAuthors: Rakesh Singh, Akshay Singh, Ajay Kumar Mahato, Rajesh Kumar, Amit K. Singh, Sundeep Kumar, Soma S. Marla, Ashok Kumar, and NK Singh Commercialization: Freely Available for non-commercial purposes.
Commercialization of Technologies
- GM Detection Technologies transferred: Five cost-efficient/rapid GMO screening technologies transferred to M/s DSS Imagetech Private Limited, New Delhi (2015)
- GMO Detection Kits:Two GM detection kits (GMO Detection Kit (35S/NOS) and FMV (GMO) Detection Kit)commercialized by the DSS Imagetech Private Limited, New Delhi(2022)
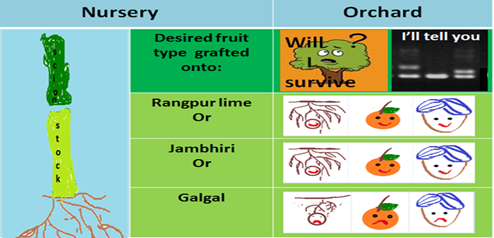
- Detection Technology: Distinction of Citrus rootstocks namely Jambhiri and Rangpur lime from Galgal using SSR based markers.
Awards/Honors
| Dr Rakesh Singh |
|
| Dr Lalit Arya |
|
| Dr Manjusha Verma |
|
| Dr Amit Kumar Singh |
|
| Dr Monika Singh |
|
| Dr. Dhammaprakash Pandhari Wankhede |
|
Major Activities
- Generation and Conservation of Genomic Resources
- Whole genome sequencing
- Database development
- Molecular systematics
- Services for DNA profiling of crop varieties
- Services for GM detection
Publications/Research Papers
2024
- Abhishek GJ, Tripathi K, SharmaSK, Parameshwari B, Gupta N, Diksha D, Ghosh, Archak S, Wankhede DP, Deepika DD, Danakumar T, KalaiponmaniK,Vignesh Muthusamy V. and Chalam V.C. (2024) Identification ofcowpea [Vigna unguiculata (L.) Walp.] germplasm accessionsresistant to yellow mosaic disease. Indian Journal of Genetics and Plant Breeding,84(2): 258-265.
- Amit KM, Neeraj B, Manjeet K, Anjan KP, Sundeep K, Prashanth B, Rajbir Y and Kiran BG (2024). Exploring the genetic diversity and population structure of an ancient hexaploid wheat species Triticum sphaerococcum using SNP markers. BMC Plant Biology, 24:1188.
- Avashthi, H., Angadi, U., Chauhan, D., Mishra, D., Sharma, A., Kumar, D., Rangan, P., Yadav, R. Sesame genomic web resource (SesameGWR): a well-annotated data resource for transcriptomic signatures of abiotic and biotic stress responses in sesame (Sesamum indicum L.). Briefings in Functional Genomics, 23(6): 828-842.
- Bairwa RK, Yadav MC, Tiwari S, Aswin M, Ellur RK and Gopalakrishnan S (2024). Aromatic rice (Oryza sativa L.) landraces of Indo-Gangetic Plains of India harbor rich genetic diversity. Crop Science 64: 1711-1728. Doi: 10.1002/csc2.21211.
- Bhardwaj, R., Gayacharan, Gawade, B. H., Pathania, P., Talukdar, A., Kumar, P., … & Singh, G. P. (2024). Identification of heat-tolerant mungbean genotypes through morpho-physiological evaluation and key gene expression analysis. Frontiers in Genetics, 15, 1482956.
- Bharti U, Gaur P, Kaur K and M Singh(2024) Tracking genetically modified (GM) rice ingredients in samples of packed rice and food products from the marketplace in India: a pilot study for regulatory compliance. Journal of Environmental Health Science and Engineering, 22(1): 263-269.
- Choudhury DR, Sharma L, Suma A, Singh GP and Singh R (2024) Exploring the genetic diversity and population structure of Bacopa monnieri (L.) using random amplified (RAPD and ISSR) and gene-targeted (SCoT and CBDP) markers. Genetic Resources and Crop Evolution, pp.1-21.
- Dhakad, A. VB Patel, SKSingh, MKVerma, GP Mishra, VBMhetre, ChavleshKumar, Rakesh Singh, S.Rajkumar, and Kishor Gaikwad (2024) Pollen biology and metaxenic studies on early maturing grape (Vitis vinifera) genotypes. The Indian Journal of Agricultural Sciences 94(10): 1094-99.
- Divya C, Dwijesh CM, Shikha M, Sushma R, Jyotika B, Sundeep K, Subhash CB, Monendra G, Neeraj B, Suphiya K (2024). Identification of hub genes associated with stripe rust disease in wheat through integrative transcriptome and gene-based association study. South African Journal of Botany 171, 583-591.
- Divya S, Neeraj B, Anita K, Dinesh KS, Anshu S, Aakash Y, Reyazul RM, Amit KS, V.K Vikas, Gyanendra PS and Sundeep K. (2024). Exploring the Genetic Architecture of Powdery Mildew Resistance in Wheat through QTL MetaAnalysis. Frontiers in Plant Sci. 10: 3389.
- Gawade, B. H., Kumari, L., Khan, Z., Saroha, A., Ujjainwal, S., Pal, D., Gayacharan., … & Wankhede, D. P. (2024). Evaluation of diverse blackgram (Vigna mungo (L.) Hepper) germplasm accessions for root-knot nematode resistance and genetic diversity study. Genetic Resources and Crop Evolution, 1-18.
- Jyoti K, RK G, Arun G, BK H, Shyam SV, Achla S, Sewa R, Gopalareddy K, Shivani S, Rakesh B, Sherry Rachel J, Sundeep K, VK Vikas, Sushil P, JC R, Ashok K, GP Singh, Kuldeep S (2024). Multi-environment analysis to unravel bread wheat core collection to identify donors for grain quality, phenology, and yield traits. Crop & Pasture Science,75, CP22340.
- Kaur S, Singh N, Sharma P, Ananthan R, Singh M, Gayacharan, SinghAK, Bhardwaj, R. (2024). Optimizing protein content prediction in rice bean (Vigna umbellata L.) by integrating near-infrared reflectance spectroscopy, MPLS, deep learning, and key wavelengths selection algorithms. Journal of Food Composition and Analysis, 135, 106655.
- Kaur V, Gomashe SS, Yadav SK, Singh D, Sheela, Chauhan SS, Kumar V, Jat B, Tayade NR, Langyan S, Kaushik N, Singh M, Kheralia M, Wankhede DP, Aravind J, Srivastava V, Gupta K, Kumar A, Singh GP (2024) Leveraging genetic resource diversity and identification of trait-enriched superior genotypes for accelerated improvement in linseed (Linumusitatissimum L.). Sci Rep. 14(1):20266. doi: 10.1038/s41598-024-71044-8
- Kondal V, Jain A, Garg M, Kumar S, Singh AK, Bhardwaj R, & Singh GP (2024). Gap derivative optimization for modeling wheat grain protein using near‐infrared transmission spectroscopy. Cereal Chemistry, 2024, 1–11.
- Kuldeep Tripathi,R K Gautam, Vijay K Verma, D P Wankhede, Rakesh Bhardwaj, S K Chaudhari and G P Singh (2024) Gadmal: An endemic pulse crop from Betul district, Madhya Pradesh, India. Indian Journal of Traditional Knowledge. 23: 3, pp 239-246DOI: 10.56042/ijtk.v23i3.1658
- KumarAjay, Tamanna Batra, Harinder Vishwakarma, Rasna Maurya, Pradeep Ruperao, Rashmi Yadav, Rajkumar Subramani, Gyanendra Pratap Singh, ParimalanRangan (2024) Genome-wide analysis of the calmodulin-binding transcription activator (CAMTA) gene family in Sesamumindicum L., and its role in abiotic stress tolerance traits. Plant Stress13: 100532.
- KumarRajesh, J. Radhamani, S. Rajkumar2024 Exploration of allelic diversity reveals a novel FAD2 (Oleate desaturase) gene in Brassica juncea. Crop Breeding and Applied Biotechnology 24(2): e47702424,
- KumarRajesh, S Rajkumar, J Radhamani (2024) Molecular Profiling of Erucic Acid Biosynthesizing FAE1 Gene of Indian Mustard Varieties. Indian Journal of Plant Genetic Resources 37(1): 20-23.
- Lone J K, Pandey R, (2024). Microgreens on the rise: Expanding our horizons from farm to fork. Heliyon, 10(4), e25870
- Manish K V, Pradeep KB, Uttam K, Ravi PS, Sundeep K, Velu G, Gurvinder SM, Karthikeyan T, Narain D, Arun KJ (2024). Genetic dissection of value-added quality traits and agronomic parameters through genome-wide association mapping in bread wheat (T. aestivum L.). Frontiers in Plant Sciences, Volume 15 – 2024.
- Maurya Rasna, Shivani Singh, Yangala Sudheer Babu, Fatima Nazish Khan, Bhagwat Nawade, Harinder Vishwakarma, Ajay Kumar, Rashmi Yadav, RadhamaniJalli, Mahalingam Angamuthu, MothilalAlagirisamy, Rajkumar Subramani, SenthilrajaGovindasamy, Ashok Kumar, Kuldeep Singh, ParimalanRangan.(2024) Molecular diversity studies and core development in sesame germplasm (sesamum indicum l.) using ssr markers. Plant Molecular Biology Reporter 1: 17
- Padhi SR, John R, Tripathi K, Wankhede DP, Joshi T, Rana JC, Riar A, Bhardwaj R (2024) A comparison of spectral preprocessing methods and their effects on nutritional traits in cowpea germplasm. Legume Science, 6:e229 https://doi.org/10.1002/leg3.229
- ParimalanRangan, Dhammaprakash P Wankhede, Rajkumar Subramani, Viswanathan Chinnusamy, Pooja Pathania, Arti Bartwal, Surendra K Malik, Mirza JaynulBaig, Anil Rai, Kuldeep Singh (2024) Rice Transcriptomics Reveal the Genetic Determinants of An In Planta Photorespiratory Bypass: a Novel Way to Increase Biomass in C3 Plants. Plant Molecular Biology Reporter 1: 10.
- Prabhu,P. Anjula Pandey, S Rajkumar, GD Harish, Chithra Pandey, Rakesh Bharadwaj (2024) Studies on Traditional Melon ‘Vellari’(Cucumismelo var. momordica L.) in Tamil Nadu, India, Indian Journal of Plant Genetic Resources 37(1): 97-104.
- Rangan, P, Furtado, A., Chinnusamy, V., and Henry, R. A multi-cell model for the C4 photosynthetic pathway in developing wheat grains based upon tissue-specific transcriptome data. BioSystems, 238: 105195.
- Rangan, P., Pradheep, K., Mahalingam, A., Mohan, J., Yadav, R., and Singh, K. Trans situ conservation strategies to conserve the extinction risk species, Sesamum prostratum Retz., a crop wild relative of sesame being endemic to coastal strand habitat: a case study. Genetic Resources and Crop Evolution., 71: 511-517.
- Rangan, Wankhede DP, Rajkumar S, Chinnusamy V, Pathania P, Bartwal A, Malik SK, Baig MJ, RaiAK, Singh K. Rice transcriptomics reveal the genetic determinants of an in planta photorespiratory bypass: a novel way to increase biomass in C3 plants. Plant Molecular Biology Reports. https://doi.org/10.1007/s11105-024-01469-y
- Roy, S., Priyanka, Sahi, A.N., Radhamani, J. and Kumar, R. (2024) High-throughput screening identifies robust root trait in Indian mustard seedlings grown under salinity stress. Genetic Resources and Crop Evolution, 71(5):1869-1882
- Ruperao P, Bajaj P, Yadav R, Angamuthu M, Subramani R, Rai V, Tiwari K, Rathore A, Singh K, Singh GP, Angadi UB, Mayes S, Rangan P. Double-digest restriction-associated DNA sequencing-based genotyping and its applications in sesame germplasm management. Plant Genome. 17(2):e20447. doi: 10.1002/tpg2.2044
- Sahu TK, Verma SK, Gayacharan, Singh NP,Joshi DC, Wankhede DP, Singh M, Bhardwaj R, Singh B, Parida SK, Chattopadhyay D, Singh GP, Singh AK (2024) Transcriptome-wide association mapping provides insights into the genetic basis and candidate genes governing flowering, maturity and seed weight in rice bean (Vigna umbellata). BMC Plant Biol 24, 379 .https://doi.org/https://pubmed.ncbi.nlm.nih.gov/38720284/
- Shanmugaraj, C., Kamil, D., Rangan, P., Singh, P.K., Shashank, P.R., Iquebal, M.A., Hussain, Z., Das, A., Gogoi, R., Nishmitha, K. Deciphering the defense response in tomato against Sclerotium rolfsii by Trichoderma asperellum strain A10 through gene expression analysis. 3 Biotech, 14: 210.
- Sharma L, Roy S, Satya P, Alam NM, Goswami T, Barman D, Bera A, Saha R, Mitra S and Mitra J (2024) Exogenous ascorbic acid application ameliorates drought stress through improvement in morpho-physiology, nutrient dynamics, stress metabolite production and antioxidant activities recovering cellulosic fibre production in jute (Corchorus olitorius L.). Industrial Crops and Products, 217:118808.
- Shivaprasad KM, Dikshit HK, Mishra GP, Sinha SK, Aski M, Kohli M, Mishra DC, Singh AK, Gupta S, Singh A, Tripathi K, Kumar RR, Kumar A, Jha GK, Agrawal S, Varshney R (2024) Delineation of loci governing an extra‐earliness trait in lentil (Lens culinarisMedik.) using the QTL‐Seq approach. Plant Biotechnology Journal. 22(10):2932-49.
- Shrinkhla Maurya, Bhumika Sharma, Kajal Thakur, Suman Rawte, Nirmala Bharti Patel, Rajani Bisen, S Rajkumar, Zenu Jha, M Sujatha (2024) In vitro double haploid production through anther culture in niger (Guizotiaabyssinica L.F. Cass)In Vitro Cellular & Developmental Biology-Plant 60(1): 50-66.
- Singh AK, Sharma L, Goswami T, Satya P, Kar G (2024) Lifecycle GHG emissions for bioethanol production from lignocellulosic biomass: potential of jute and kenaf feedstock from an Indian perspective. Current Science,127(9):1076-1082
- Singh M, Pal D, Aminedi R and AK Singh (2024) Multiplex real-time loop-mediated isothermal amplification (LAMP) based on the annealing curve analysis: toward an on-site multiplex detection of transgenic sequences in seeds and food products. Journal of Agricultural anfFood Chemistry72 (31): 17658-17665.
- Singh, A. Avantika Maurya, Subramani Rajkumar, Amit Kumar Singh, Rakesh Bhardwaj,Surinder Kumar Kaushik, Sandeep Kumar, Kuldeep Singh, Gyanendra Pratap Singh and Rakesh Singh (2024) Comparative analysis of five amaranthaceae species reveals a large amount of repeat content. Plants 13(824):1-26.
- Suman Dutta, Rashmi Chhabra, Vignesh Muthusamy, Nisrita Gain, Rajkumar Subramani, Konsam Sarika, Elangbam L Devi, Jayanthi Madhavan, Rajkumar U Zunjare, Firoz Hossain (2024) Allelic variation and haplotype diversity of Matrilineal (MTL) gene governing in vivo maternal haploid induction in maize. Physiology and Molecular Biology of Plants 1-16.
- Verma, J.; Gore, P.G.; Kumari, J.; Wankhede, D.P.; Jacob, S.R.; Thirumani Venkatesh, A.K.; Nair, R.M.; Tripathi, K. (2024) Exploring genetic diversity in black gram (Vigna mungo (L.) Hepper) for pre-harvest sprouting tolerance. Agronomy, 14, 197. https://doi.org/10.3390/ agronomy14010197.
- Vishwakarma, H., Sharma, S., Panzade, K.P., Kharate, P.S., Kumar, A., Singh, N., Avashthi, H., Rangan, P., Singh, A.K., Singh, A., Angadi, U.B., Siddique, K.H.M., Singh, K., Singh, G.P., Pandey, R., and Yadav, R. Genome-wide analysis of the class III peroxidase gene family in sesame and SiPRXs gene validation by expression analysis under drought stress. Plant Stress, 11: 100367
- Yadav AK, CK Singh, DP Wankhede, RK Kalia, S Pradhan, S Ujjainwal, TK Sahu, NS Nathawat, RK Kakani, R Rani, R Kumar, R Pathak, R Singh, GP Singh, AK Singh (2024) Combined genome‑wide association study and expression analysis unravels candidate genes associated with seed weight in moth bean [Vigna aconitifolia (Jacq.) Marechal].Journal of Plant Growth Regulation. https://doi.org/10.1007/s00344-024-11517-2
- Yadav S, Kumari R, Rangan P, Gaikwad AB. (2024) Variability in genome size of Trigonellafoenum-graecum, Trigonellacorniculata and Trigonellacaerulea as estimated by flow cytometry indicates complex evolutionary history of fenugreek. MolecularBiology Reports, 51, 489. https://doi.org/10.1007/s11033-024-09417-5
- Yadav S, Yadava YK, Meena S, Kalwan G, Bharadwaj C, Paul V, Kansal R, Gaikwad K, Jain PK. (2024) Novel insights into drought-induced regulation of ribosomal genes through DNA methylation in chickpea. International Journal of Biol Macromolecules. 266(Pt 2):131380.
2023
- Aswin M, Yadav MC, Gopalakrishnan S, Tiwari S and Mondal TK (2023) Phenotypic characterization of crop genetic resources of AA-genome species in rice (Oryza spp.). Indian Journal of Plant Genetic Resources 36(3) 402-414. doi: 10.61949/0976-1926.2023.v36i03.09.
- Aswin M, Yadav MC, Tiwari S, Bairwa RK, Gopalakrishnan S, Rana MK, Singh R and Mondal TK (2023) Population structure and genetic differentiation analyses reveal high level of diversity and allelic richness in crop wild relatives of AA-genome species of rice (Oryza sativa L.) in India. Journal of Applied Genetics 64: 645-666. doi: 10.1007/s13353-023-00787-3.
- Bairwa, RK, Yadav MC, Gopalakrishnan S, Kushwaha AK and Joshi, MA (2023) Morphological and molecular analyses of grain traits in aromatic rice landrace accessions from Indo-Gangetic plain region of India. Indian Journal of Plant Genetic Resources 36 (2): 290-300. doi: 10.5958/0976-1926.2023.00036.2.11.
- Bansal S, Sharma MK, Singh S, Joshi P., Pathania P, Malhotra EV, S Rajkumar, Misra P (2023). Histological and molecular insights in to in vitro regeneration pattern of Xanthosomas agittifolium. Sci. Rep 13, 5806. https://doi.org/10.1038/s41598-023-33064-8
- Bhutia ND, Sureja AK, Verma M, Gopala Krishnan S, Arya L, Bhardwaj R (2023) Inheritance and molecular mapping of solitary/cluster fruit-bearing habit in Luffa. Mol. Genet. Genom. 1-12. https://doi.org/10.1007/s00438-023-02034-0
- Chandra T, S Jaiswal, MA Iquebal, R Singh, RK Gautam, A Rai, D Kumar (2023) Revitalizing miRNAs mediated agronomical advantageous traits improvement in rice,Plant Physiology and Biochemistry,107933(IF: 6.50)
- Choudhury DR, R Kumar, A Maurya, DP Semwal, RS Rathi, RK Gautam, AK Trivedi, SK Bishnoi, SP Ahlawat, Kuldeep Singh, NK Singh, and R Singh*(2023)SSR and SNP marker-based investigation of Indian rice land-races in relation to their genetic diversity, population structure, and geographical isolation. Agriculture.13(4), 823 (IF: 3.40)
- Das P, Grover M, Chauhan D, Mishra DC, Kumar S, Chaturvedi KK, Bhardwaj SC, Singh AK, Rai A (2023) Comparative transcriptome analysis of wheat isogenic lines provides insights into genes and pathways associated with stripe rust resistance. Indian Journal of Genetics and Plant Breeding 83 (1), 52-58. https://doi.org/10.31742/ISGPB.83.1.7
- Deepika DD, SR Padhi, PG Gore, K Tripathi, AK Katral, KR Chandora, GJ Abhishek, V Kondal, R Singh, R Bharadwaj, KC Bhatt, JC Rana and A Riar (2023) Nutritional Potential of Adzuki Bean Germplasm and Mining Nutri-Dense Accessions through Multivariate Analysis. Foods 12, 4159.(IF: 5.20)
- Francis A, Singh NP, Singh M, Sharma P, Gayacharan, Kumar D, Basu U, Bajaj D, Varshney N, Joshi DC, Semwal DP, Tyagi V, Wankhede D, Bharadwaj R, Singh AK, Parida SK and Chattopadhyay D (2023) The ricebean genome provides insight into Vigna genome evolution and facilitates genetic enhancement. Plant Biotechnol. J., https://doi.org/10.1111/pbi.14075.
- Gaikwad AB, Kaila T, Maurya A, Kumari R, Rangan P, Wankhede DP and Bhat KV (2023) The chloroplast genome of black pepper (Piper nigrum L.) and its comparative analysis with related Piper species. Frontiers in Plant Sciences 13:1095781. doi: 10.3389/fpls.2022.1095781.
- Gaikwad AB, Kumari R , Yadav S, Rangan P, Wankhede DP and Bhat KV (2023). Small Cardamom genome: Development and utilization of microsatellite markers from a draft genome sequence of Elettaria cardamomum Maton. Front. Plant Sci, 14 – 2023. https://doi.org/10.3389/fpls.2023.1161499
- Htwe CSS, Rajkumar S, Pathania P, Agrawal A (2023) Transcriptome profiling during sequential stages of cryopreservation in banana (Musa AAA cv Borjahaji) shoot meristem. Plants , 12, 1165. https://doi.org/10.3390/plants12051165
- John R, Bollinedi H, Jeyaseelan C, Padhi SR, Neha, Nath D, Singh R, Ahlawat SP, Bhardwaj R, Rana JC (2023) Mining nutri-dense accessions from rice landraces of Assam, India. Heliyon:e17524(IF: 3.77)
- Kabade, P; S Dixit, UM Singh, S Alam, S Bhosale, S Kumar; SK Singh, J Badri, NR Gopala Varma, RNadimpalli, S Chetia, RSingh, S Pradhan, S Banerjee, R Deshmukh, S Singh, S Kalia, TR Sharma, S Singh, H Bhardwaj, A Kohli, A Kumar, P Sinha, V Singh (2023) SpeedFlower: a comprehensive speed breeding protocol for indica and japonica rice. Plant Biotechnology Journal. 21 (12), 1-16. DOI: 10.1111/pbi.14245 (IF: 13.8)
- Kalwan G, Gill SS, Priyadarshini P ,Gill R, Yadava YK, Yadav S, Baruah P, Agarwala N, Gaikwad K, Jain PK (2023) Approaches for identification and analysis of plant circular RNAs and their role in stress responses. Environmental and Experimental Botany, 205: 105099. https://doi.org/10.1016/j.envexpbot.2022.105099
- Kalwan G, Priyadarshini P, Kumar K, Yadava YK, Yadav S, Kohli D, Gill SS, Gaikwad K, Hegde V and Jain PK (2023) Genome wide identification and characterization of the amino acid transporter (AAT) genes regulating seed protein content in chickpea (Cicer arietinum L.). Int J Biol Macromol, 252: 126324. DOI: 10.1016/j.ijbiomac.2023.126324
- Kaur V, Singh M, Wankhede DP, Gupta K, Langyan S, Aravind J, Thangavel B, Yadav SK, Kalia S, Singh K and Kumar A (2023) Diversity of Linum genetic resources in global genebanks: from agro-morphological characterisation to novel genomic technologies – a review. Front. Nutr. 10:1165580. DOI: 10.3389/fnut.2023.1165580
- Kumar GP, Pathania P, Goyal N, Gupta N, Parimalan R, Radhamani J, Gomashe SS, Kadirvel P, Rajkumar S (2023) Genetic diversity and population structure analysis to construct a core collection from safflower (Carthamus tinctorius L.) germplasm through SSR markers. Agriculture 13, 836. https://doi.org/10.3390/agriculture13040836
- Kumar S, Saini DK, Jan F, Jan S, Tahir M, Djalovic I, Latkovic D, Khan MA, Kumar S, Vikas VK, Kumar U, Kumar S, Dhaka NS, Dhankher OP, Rustgi S and Mir RR (2023) Comprehensive meta-QTL analysis for dissecting the genetic architecture of stripe rust resistance in bread wheat.BMC Genomics, 24:259. DOI: 10.1186/s12864-023-09336-y.
- Kumari J, Lakhwani D, Jakhar P, Sharma S, Tiwari S, Mittal S, Avashthi H, Shekhawat N, Singh K, Mishra KK, Singh R, Yadav MC, Singh GP, Singh AK (2023). Association mapping reveals novel genes and genomic regions controlling grain size architecture in mini core accessions of Indian National Genebank wheat germplasm collection. Frontiers in Plant Science, 28; 14:1148658. https://doi.org/10.3389/fpls.2023.1148658
- Kumari M, SR Padhi, SK Chourey, V Kondal, SS Thakare, A Negi, V Gupta, M Arya, JK Yasin, R Singh, C Bharadwaj; A Kumar; R Bharadwaj; KC Bhatt; JC Rana, T Joshi; A Riar (2023) Unveiling diversity for quality traits in Horsegram {Macrotyloma uniflorum (Lam.) Verdc.} landraces: A forgotten nutraceutical pulse crop of ancient India. Plants 12, 3803.
- Mahendran A, MC Yadav, S Tiwari, RK Bairwa, SGKrishnan, MK Rana,R Singh and TK Mondal (2023) Population Structure and Genetic Differentiation Analyses RevealHigh Level of Diversity and Allelic Richness in Crop Wild Relatives of AA-Genome Species of Rice (Oryza sativa L.) in India. Journal of Applied Genetics https://doi.org/10.1007/s13353-023-00787-3.(IF: 2.4)
- Manju, Yadav SK, Wankhede DP, Saroha A, Jacob SR, Pandey R, Sharma AD, Sheela, Manoj Kumar, Kaur V. (2023) Screening of barley germplasm for drought tolerance based on root architecture, agronomic traits and identification of novel allelic variants of HVA1. Journal of Agronomy and Crop Science. https://doi.org/10.1111/jac.12650
- Mir ZA, Chandra T, Saharan A, Budhlakoti N, Mishra DC, Saharan MS, Mir RR, Singh AK, Sharma S, Vikas VK, Kumar S (2023).Recent advances on genome-wide association studies (GWAS) and genomic selection (GS); prospects for Fusarium head blight research in Durum wheat. Molecular Biology Reports, DOI:10.1007/s11033-023-08309-4.
- Mir ZA, Chauhan D, Pradhan AK, Srivastava V, Sharma D, Budhlakoti N, Mishra DC, Jadon V, Sahu TK, Grover M, Gangwar OP, Kumar S, Bhardwaj SC, Padaria JC, Singh AK, Rai A, Singh GP andKumar S (2023) Comparative transcriptome profiling of near isogenic lines PBW343 and FLW29 to unravel defense related genes and pathways contributing to stripe rust resistance in wheat. Functional & Integrative Genomics.doi.org/10.1007/s10142-023-01104-1.
- Nagar A, Gowthami R, Sureja AK, Munshi AD, Verma M, Singh AK, Mallick N, Singh J, Chander S, Shankar M, Pathania P and Rajkumar S (2023) Simple cryopreservation protocol for Luffa pollen: enhancing breeding efficiency. Front. Plant Sci. 14:1268726. doi: 10.3389/fpls.2023.1268726
- Natarajan RB, Pathania P, Singh H, Agrawal A, Subramani R (2023) A flow cytometry-based assessment of the genomic size and ploidy level of wild Musa species in India. Plants, 12, 3605. https://doi.org/ 10.3390/plants12203605.
- Nishmitha K, R Singh, SC Dubey, J Akhtar, K Tripathi, and D Kamil (2023) Resistance screening and in-silico characterization of cloned novel RGA from multi-race resistant Lentil germplasm against Fusarium wilt (Fusarium oxysporum f. sp. Len lentils. Frontiers in Plant Science. 14. 1147220.
- Nishmitha K., R Singh, J Akthar, BM Baishaland D Kamil (2023) Expression profiling and characterization of key RGA involved in Lentil Fusarium wilt Race 5 resistance. World Journal of Microbiology and Biotechnology.39:306.
- Patidar A, Yadav MC, Kumari J, Tiwari S, Chawla G and Paul V (2023) Identification of Climate-Smart Bread Wheat Germplasm Lines with Enhanced Adaptation to Global Warming. Plants 12(15):2851. doi: 10.3390/plants 12152851.
- Pillai MA, E M, Michael antonysamy Tamilarasi AP, Selvaraj R, Devadhasan S, Yasin JK, Jegadeesan S. (2023) Genotype‐by‐environmental interaction effects on earliness, semi‐dwarfism, and yield of rice (Oryza sativa L.) mutants in Tamil Nadu, India. Crop Science. 63(6):3402-15.
- Pradhan AK, Budhlakoti N, Mishra DC, Prasad P, Bhardwaj SC, Sareen S, Sivasamy M, Jayaprakash P, Geetha M, Nisha R, Shajitha P, Peter J, Kaur A, Kaur S, Vikas VK, Singh K, and Kumar S* (2023)Identification of Novel QTLs/Defense Genes in Spring Wheat Germplasm Panel for Seedling and Adult Plant Resistance to Stem Rust and Their Validation Through KASP Marker Assays. Plant Disease. doi.org/10.1094/PDIS-09-22-2242-RE.
- Prathyusha VB, Swathi E, Divya D, Reddy BVB, Bentur JS, Chalam VC, Wankhede DP, Singh K, Anitha K. (2023) Field and agroinoculation screening of national collection of urd bean (Vigna mungo) germplasm accessions for new sources of mung bean yellow mosaic virus (MYMV) resistance. 3 Biotech. 2023 Jun;13(6):194. DOI: 10.1007/s13205-023-03610-2
- R Tyagi, R Bhardwaj, P Suneja, AK Sureja, AD Munshi, L Arya, A Riar, M Verma (2023) Harnessing sponge gourd: an alternative source of oil and protein for nutritional security. Front. Nutr. 10, 1158424. https://doi.org/10.3389/fnut.2023.1158424
- Ramya K R, R. Gupta, M Mishra, A. Pandey, K Tripathi, K C Bhatt, R Singh (2023) A modified DNA isolation protocol for high-quality DNA and long-term storability in Grass pea (Lathyrus sativus L.). Indian Journal of Genetics and Plant Breeding 83(4):602-604.
- Roy S, Priyanka, Sahi AN, Radhamani J and Kumar R (2023) High-throughput screening identifies robust root trait in Indian mustard seedlings grown under salinity stress. Genetic Resources and Crop Evolution, pp.1-14. (https://doi.org/10.1007/s10722-023-01743-0)
- Ruperao P, P Bajaj, S Rajkumar, R Yadav, VBR Lachagari, SP Lekkala, A Rathore, S Archak, UB Angadi, R Singh, K Singh, S Mayes, P Rangan (2023) A pilot-scale comparison between single and double-digest RAD markers generated using GBS strategy in sesame (Sesamum indicum L.). Plosone 18 (6), e0286599. https://doi.org/10.1371/journal.pone.0286599
- S Mahapatra, AK Sureja, TK Behera, R Bhardwaj, M Verma (2023) Variability in antioxidant capacity and some mineral nutrients among ninety-one Indian accessions of bottle gourd [Lagenaria siceraria (Molina) Standl.] S. Afr. J. Bot. 152, 50-62. https://doi.org/10.1016/j.sajb.2022.11.040
- Saroha A, Gomashe SS, Kaur V, Pal D,Ujjainwal S, Aravind J, Singh M, Rajkumar S,Singh K, Kumar A and Wankhede DP* (2023) Genetic dissection of thousand-seedweight in linseed (Linum usitatissimum L.)using multi-locus genome-wide association study. Front. Plant Sci. 14:1166728.doi: 10.3389/fpls.2023.1166728. https://doi.org/10.3389/fpls.2023.1166728
- Sharma D, Kumari A, Sharma P, Singh A, Sharma A, Mir ZA, Kumar U, Jan S, M. Parthiban, Mir RR, Bhati P, Pradhan AK, Yadav A, Mishra DC, Budhlakoti N, Yadav MC, Gaikwad KB, Singh AK, Singh GP and Kumar S (2023). Meta-QTL analysis in wheat: progress, challenges and opportunities. Theoretical and Applied Genetics 136:247 https://doi.org/10.1007/s00122-023-04490-z.
- Sharma P, Goudar G, Chandragiri AK, Ananthan R, Subhash K, Chauhan A, Longvah T, Singh M, Bhardwaj R, Parida SK, Singh AK, Gayacharan, Chattopadhyay D (2023) Assessment of diversity in anti-nutrient profile, resistant starch, minerals and carbohydrate components in different ricebean (Vignaumbellata) accessions. Food Chemistry. 30; 405:134835. https://doi.org/10.1016/j.foodchem.2022.134835
- Shruti, A Shukla, SS Rahman, P Suneja, R Yadav, Z Hussain, R Singh, SK Yadav, JC Rana, S Yadav, and R Bhardwaj (2023) Developing an NIRS Prediction Model for Oil, Protein, Amino Acids and Fatty Acids in Amaranth and Buckwheat.Agriculture 13(2), 469. (IF: 3.40)
- Singh A, Mahato AK, Maurya A, Rajkumar S, Singh AK, Bhardwaj R, Kaushik SK, Kumar S, Gupta V, Singh K and Singh R (2023) Amaranth Genomic Resource Database: an integrated database resource of Amaranth genes and genomics. Frontiers in Plant Science. 14:1203855. doi: 10.3389/fpls.2023.1203855
- Singh M, A Paliwal, K Kaur, P Palit and G Randhawa (2023). Development and utilization of analytical methods for rapid GM detection in processed food products: a case study for regulatory requirement. Journal of Plant Biochemistry and Biotechnology, 32: 511–524. https://doi.org/10.1007/s13562-023-00832-6
- Singh M, Palit P, Kaur K, Aminedi R, Paliwal A and Randhawa G (2023) Loop-mediated isothermal amplification targeting proteinase inhibitor II terminator sequence: an efficient approach for screening of genetically modified crops. European Food Research and Technology. 249, 2311-2319. https://doi.org/10.1007/s00217-023-04294-x
- Singh M, Papazova N, Aminedi R, Fraiture M-A, Roosens N and Randhawa G (2023). Approaches to check unauthorized genetically modified events in supply chain: Challenges and solutions in the Indian context. JSFA Reports, 3 (5): 184-195. https://doi.org/10.1002/jsf2.115
- Singh R, A Singh, AK Mahato, R Paliwal, G Tiwari, and A Kumar (2023) De Novo Transcriptome Profiling for the Generation and Validation of Microsatellite Markers, Transcription Factors, and Database Development for Andrographis paniculata. International Journal of Molecular Sciences. Int. J. Mol. Sci. 24,9212.
- Singh VK, Chaturvedi D, Pundir S, Kumar D, Sharma R, Kumar S, Sharma Sand Sharma S (2023) GWAS scans of cereal cyst nematode (Heteroderaavenae) resistance in Indian wheat germplasm.Molecular Genetics and Genomics.DOI: 10.1007/s00438-023-01996-5.
- Singh SK, Deo S, Chaudhary SB, Singh K and Rana MK (2023). Genetic Relationships and Population Structure among Maize (Zea mays L.) Landraces as Revealed by Simple Sequence Repeat Markers. Indian J. Plant Genetic Resources. 36(2), 216-226.DOI: 10.61949/0976-1926.2023.v36i02.03
- Srivastav M, N Radadiya, S Ramachandra, P Jayswal, N Singh, S Singh, AK Mahato, G Tandon, A Gupta, R Devi, SH Subrayagowda, G Kumar, P Prakash, S Singh, N Sharma, A. Nagaraja, A Kar, SG Rudra, S Sethi, S Jaiswal, MA Iquebal, R Singh, SK Singh and NK Singh (2023) High resolution mapping of QTLs for fruit color and firmness in Amrapali/Sensation mango hybrids. Frontiers in Plant Science.14:1135285
- Upadhyay D, Budhlakoti N, Kumari J, Chaudhary N, Padaria JC, Sareen S and Kumar S (2023) In silico characterization of drought stress related WRKY2 transcription factor in wheat crop (Triticum aestivum L.):study of its physico chemical properties and structuraldynamics. Genetic Resources and Crop Evolution. DOI: https://doi.org/10.1007/s10722-023-01706-5.
- Upadhyay D, Neeraj Budhlakoti N, Mishra DC, Kumari J, Gahlaut V, Nidhee Chaudhary N, Padaria JC, Sareen S and Kumar S (2023) Characterization of stress-induced changes in morphological, physiological and biochemical properties of Indian bread wheat (Triticum aestivum L.) under deficit irrigation. Genetic Resources & Crop Evolution.DOI: https://doi.org/10.1007/s10722-023-01693-7.
- Vats G, D Das, R Gupta, ASingh, A Maurya, S. Rajkumar, AK Singh, R Bhardwaj, S Kumar, SK Kaushik, V Gupta, K Singh and R Singh (2023)Validation of Genome-wide SSR markers developed for genetic diversity and population structure study in Grain Amaranth (Amaranthus hypochondriacus).Agriculture 13(2),431. https://doi.org/10.3390/agriculture13020431
- Vinay ND, H Matsumura, AD Munshi, RK Ellur, V Chinnusamy, A Singh, MA Iquebal, S Jaiswal, GS Jat, I Panigrahi, AB Gaikwad, AR Rao, SS Dey and TK Behera (2023). Molecular mapping of genomic regions and identification of possible candidate genes associated with gynoecious sex expression in bitter gourd. Frontiers in Plant Science 14:1071648. DOI 10.3389/fpls.2023.1071648
- Yadav AK, Singh CK, Kalia RK, Mittal S, Wankhede DP, Kakani RK, Ujjainwal S, Saroha A, Nathawat NS, Rani R, Panchariya P, Choudhary M, Solanki K, Chaturvedi KK, Archak S, Singh S, Singh GP Singh AK (2023) Genetic diversity, population structure, and genome-wide association study for the flowering trait in a diverse panel of 428 moth bean (Vigna aconitifolia) accessions using genotyping by sequencing. BMC Plant Biology. 23(1):1-7.https://doi.org/10.1186/s12870-023-04215-w
- Yadav S, Kalwan G, Meena S, Gill SS, Yadava YK, Gaikwad K and Jain PK (2023). Unravelling the due importance of pseudogenes and their resurrection in plants. Plant Physiol Biochem, 203:108062. https://doi.org/10.1016/j.plaphy.2023.108062
- Yadav S, Tyagi A, Kumari R, Srivastava H, Rangan P, Wankhede DP and Gaikwad AB (2023). Comparative transcriptome profiling of fruit tissue provides novel insights into piperine biosynthesis in black pepper (Piper nigrum L.). Scientia Horticulturae, 322, 112451. https://doi.org/10.1016/j.scienta.2023.112451
- Yadav S, Yadava YK, Meena S, Singh L, Kansal R, Grover M, Nimmy MS, Bharadwaj C, Paul V, Gaikwad K, Jain PK (2023). The SPL transcription factor genes are potential targets for epigenetic regulation in response to drought stress in chickpea (C. arietinum L.). Mol Biol Rep, 50(6):5509-5517. DOI: 10.1007/s11033-023-08347-y
- Yadava YK, Chaudhary P, Yadav S et al (2023). Genetic mapping of quantitative trait loci associated with drought tolerance in chickpea (Cicer arietinum L.). Sci Rep, 13, 17623. https://doi.org/10.1038/s41598-023-44990-y
2022
- Adhikari S, Kumari J, Jacob SR, Prasad P, Gangwar OP, Lata C, Thakur R, Singh AK, Bansal R, Kumar S, Bhardwaj SC and Kumar S (2022) Landraces-potential treasure for sustainable wheat improvement.Genetic Resources & Crop Evolution,69:499–523.DOI: 10.1007/s10722-021-01310-5.
- Arya L, Narayanan RK, Kak A, Pandey CD, Verma M, Gupta V (2022) ISSR marker based genetic diversity in Morinda for its enhanced collection, conservation and utilization. Genet Resour Crop Evol 69:1585–1593. https://doi.org/10.1007/s10722-021-01321-2
- Bansal S, A Kumar; AA Lone; MH Khan; EV Malhotra; R Singh(2022) Development of novel Genome-wide simple sequence repeats (SSR) markers in Buniumpersicum.Industrial Crops and Products.178. 114625. (IF: 6.45)
- Bartwal A, John R, Padhi SR, Suneja P, Bhardwaj R, Gayacharan, Wankhede DP, Archak S. (2023) NIR spectra processing for developing efficient protein prediction Model in mungbean. Journal of Food Composition and Analysis, 116. 105087, https://doi.org/10.1016/j.jfca.2022.105087. (Impact Factor: 4.52).
- Chaurasia S, Kumar A, Singh AK. Comprehensive Evaluation of Morpho-Physiological and Ionic Traits in Wheat (Triticumaestivum) Genotypes under Salinity Stress. Agriculture. 2022; 12(11):1765. https://doi.org/10.3390/agriculture12111765
- Chongtham SK, Devi EL, Samantara K, Yasin JK, Wani SH, Mukherjee S, Razzaq A, Bhupenchandra I, Jat AL, Singh LK, Kumar A. (2022) Orphan legumes: harnessing their potential for food, nutritional and health security through genetic approaches. 256(2):24.
- Gangappa ND, C Singh, MK Verma, M Thakre, AMV Sevanthi, R Singh, M Srivastav, Raghunandan K, C Anusha, V Yadav, A Nagaraja (2022) Assessing the genetic diversity of guava germplasm characterized by morpho-biochemical traits. Frontiers in Nutrition. 9:1017680(IF: 6.59)
- Gautam, RK, Langyan S, S. Vimla Devi, Singh R, Semwal D P, Ali S, Mangat G S, Sarkar S, Bagchi TB, Roy S, Senguttuvel P, Bhuvaneswari S, Chetia S, Tripathi K, Harish GD, Kumar A, Singh K (2022) Genetic resources of sticky rice in India: status and prospects. Genetic Resourcesand Crop Evolution. 10.1007/s10722-022-01479-3 (IF: 1.88
- Gore PG, V Gupta, R Singh, K Tripathi,R Kumar, G Kumari, Latha M, HK Dikshit, Kamala V, A Pandey, N Singh, KV Bhat, RM Nairan,d APratap (2022) Understanding the genetic diversity of an underutilized Indian legume, Vigna stipulacea (Lam.) Kuntz., using morphological traits and microsatellite markers and identification of trait-specific germplasm.PLoS ONE 17(1): e0262634. (IF: 3.75)
- Gupta OP, Singh AK, Singh A, Singh GP, Bansal KC, Datta SK (2022) Wheat Biofortification: utilizing natural genetic diversity, genome-wide association mapping, genomic selection, and genome editing technologies. Front Nutr. 9:826131. DOI:3389/fnut.2022.826131
- Jain P, Singh A, Iquebal MA, Jaiswal S, Kumar S, Kumar D and Rai A (2022) Genome-Wide Analysis and Evolutionary Perspective of the Cytokinin Dehydrogenase Gene Family in Wheat (Triticumaestivum).Frontiers in Genetics 13:931659.DOI: 10.3389/fgene.2022.931659.
- John, Racheal, R. Bhardwaj,C.Jeyaseelan, H.Bollinedi, N. Singh, Harish G.D., R Singh, D. Nath, M. Arya, D. Sharma, S. Singh, J. John K,M. Latha, J.C. Rana,S.P. Ahlawat, A. Kumar (2022) Germplasm Variability Assisted NIRS Chemometrics to Develop Multi-Trait Robust Prediction Models in Rice. Frontiers in Nutrition. 2022. 946255. doi: 10.3389/nut.2022.946255 (IF: 6.59)
- Korattukudy CA, Samy AP, Rengasamy K, Devadhasan S, Shunmugavel S, Yasin JK, Madhavan AP. (2022) Genetic diversity analysis in blackgram (Vigna mungo) genotypes using microsatellite markers for resistance to Yellow mosaic virus. Plant Protection Science, 58(2), 110-124.
- Kumar R, C Kumar, R Jain, A Maurya, A Kumar, A Kumari, R Singh (2022)Molecular cloning and in-silico characterization of NAC86 of Kalmegh (Andrographis paniculata). Indian Journal of Horticulture 79(1):9-14.
- Kumar S, Pradhan AK, Kumar U, Dhillon GS, Kaur S, Budhlakoti N, Mishra DC, Singh AK, Singh R, Kumari J, Kumaran VV, Mishra VK, Bhati PK, Das S, Chand R, Singh K andKumar S(2022)Validation of Novel spot blotch disease resistance alleles identified in unexplored wheat (Triticumaestivum) germplasm lines through KASP markers. BMC Plant Biology. DOI: 10.1186/s12870-022-04013-w.
- Kumari J, Mahatman KK, Sharma S, Singh AK, Adhikari S, Bansal R, Kaur V, Kumar S and Yadav MC (2022) Recent advances in different omics mechanism for drought stress tolerance in rice. Russian Journal of Plant Physiology, 69:18. doi: 1134/S1021443722010095.
- Kumari S, Wankhede, D.P.; Murmu, S.; Maurya, R.; Jaiswal, S.; Rai, A.; Archak, S. (2022) Genome-Wide Identification and Characterization of Trihelix Gene Family in Asian and African Vigna Species. Agriculture, 12, 2172. https://doi.org/10.3390/agriculture12122172. (Impact Factor: 3.4) (*Corresponding author)
- Mhetre, Vishal B., V.B. Patel, S.K. Singh, Gyan P. Mishra, M.K. Verma, Chavlesh Kumar, Anil Dahuja, Sanjeev Kumar, R Singh and M. Wasim Siddiqui (2022) Secondary metabolites and transcriptomic analyses reveal the pathways influencing berry qualities in grapevine cv. Flame Seedless treated with bioregulators under the hot subtropical region. Food Chemistry: Molecular Sciences. 5:100116.(IF: 3.30)
- Padhi SR, Bartwal A, John R, Tripathi K, Gupta K, Wankhede DP, Mishra GP, Kumar S, Archak S and Bhardwaj R (2022) Evaluation and Multivariate Analysis of Cowpea [Vigna unguiculata (L.) Walp] Germplasm for Selected Nutrients—Mining for Nutri-Dense Accessions. Frontiers in Sustainable Food Systems. 6:888041. doi: 10.3389/fsufs.2022.888041
- Padhi SR, John R, Bartwal A, Tripathi K, Gupta K, Wankhede DP, Mishra GP, Kumar S, Rana JC, Riar A and Bhardwaj R (2022) Development and optimization of NIRS prediction models for simultaneous multi-trait assessment in diverse cowpea germplasm. Frontiers in Nutrition. 9:1001551. doi: 10.3389/fnut.2022.1001551.
- Paliwal R, R Singh, DR Choudhury, G Tiwari, A Kumar, KC Bhatt and Rita Singh (2022) Molecular Characterization of Tinospora cordifolia (Willd.) Miers Using Novel g-SSR Markers and Their Comparison with EST-SSR and SCoT Markers for Genetic Diversity Study. Genes. 13. 2042
- Pandey A, V Devi, S Nivedhitha, RK Pamarthi, R Gupta, R Singh, SP Ahlawat and K Singh (2022) Towards Strengthening the National Herbarium of Cultivated Plants with Rice Landrace Diversity. Indian J. Plant Genet. Resour. 35(1): 11-20. (IF: 0.00)
- Panigrahi, S.K., K. Tripathi, Singh,R.Kumar, P. Sanghamitra, D.P.Wankhede, N. Singh, K. K. Dubey and K.Gupta (2022). Evaluation of black gram (Vigna mungo) gene pool against Callosobruchus maculatus and diversity analysis inter se. The Indian Journal of Agricultural Sciences. 92(7):915–919. https://doi.org/10.56093/ijas.v92i7.122390
- Prasad G, Mittal S, Kumar A, Chauhan D, Sahu TK, Kumar S, Singh R, Yadav MC and Singh AK (2022) Transcriptome analysis of bread wheat genotype KRL3-4 provides a new insight into regulatory mechanisms associated with sodicity (high pH) tolerance. Frontiers in Genetics. 12:782366. doi: 3389/fgene. 2021.782366.
- Puneeth, P.V., Lata, S., Yadav, R.K., Wankhede D. P. et al. Exploring the genetic diversity using CAAT box-derived polymorphism (CBDP) and start codon targeted (SCoT) markers in cultivated and wild species of okra (Abelmoschus esculentus (L.) Moench). Genetic Resources and Crop Evolution (2022). https://doi.org/10.1007/s10722-022-01458-8 (Impact Factor: 1.87).
- Raghunandan K, Tanwar J, Patil SN, Chandra AK, Tyagi S, Agarwal P, Mallick N, Murukan N, Kumari J, Sahu TK, Jacob SR, Kumar A, Yadav S, Nyamgoud S, Vinod, Singh AK*, Jha SK. Identification of Novel Broad-Spectrum Leaf Rust Resistance Sources from Khapli Wheat Landraces. Plants. 2022; 11(15):1965. https://doi.org/10.3390/plants11151965
- Ram C, Danish S, Kesawat MS, Panwar BS, Verma M, Arya L, Yadav S, Sharma V (2022). Genome-wide comprehensive characterization and expression analysis of TLP gene family revealed its responses to hormonal and abiotic stresses in watermelon (Citrullus lanatus). Gene, 844:146818. DOI:1016/j.gene.2022.146818
- Rangan P, Wankhede DP, Subramani R, et al. 2022 Evolution of an intermediate C4 photosynthesis in the non-foliar tissues of the Poaceae. Photosynthesis Research. DOI: 10.1007/s11120-022-00926-7.
- Sahu TK, Singh AK, Mittal S, Jha SK, Kumar S, Jacob SR and Singh K (2022) G-DIRT: a web server for the identification and removal of duplicate germplasms based on identity-by-state analysis using single nucleotide polymorphism genotyping data. Briefings in Bioinformatics.DOI:10.1093/bib/bbac348.
- Saroha A, Pal D, Gomashe SS, Akash, Kaur V, Ujjainwal S, Rajkumar S, Aravind J, Radhamani J, Kumar R, Chand D, Sengupta A and Wankhede DP (2022) Identification of QTNs Associated with Flowering Time, Maturity, and Plant Height Traits in Linum usitatissimum L. Using Genome-Wide Association Study. Frontiers in Genetics. 13:811924. https://doi:10.3389/fgene.2022.811924.
- Saroha, A., Pal, D., Kaur, V. Kumar S, Bartwal A, J. Aravind, J. Radhamani, S. Rajkumar, Kumar R, Gomashe SS, Sengupta A, Wankhede DP (2022) Agro-morphological variability and genetic diversity in linseed (Linum usitatissimum L.) germplasm accessions with emphasis on flowering and maturity time. Genetic Resources and Crop Evolution https://doi.org/10.1007/s10722-021-01231-3
- Sendhil R, Nyika J, Yadav S, Mackolil J, G RP, Workie E, Ragupathy R, Ramasundaram P (2022). Genetically modified foods: bibliometric analysis on consumer perception and preference. GM Crops Food, 13(1): 65-85. doi: 1080/21645698.2022.2038525
- Shivran M, NSharma, AK Dubey, SK Singh, N Sharma, RMSharma, N Singh and R Singh (2022) Scion–Rootstock Relationship: Molecular Mechanism andQuality Fruit Production. 12,2036.
- Shweta Kumari, Ujjainwal S, Singh N., Archak S, Wankhede D.P. (2022). Development of Genic Simple Sequence Repeat Markers as Novel Genomic Resources in Dolichos Bean (Lablab purpureus L.) Indian J. Plant Genet. Resour. 35(1): 80–84 (2022) DOI 10.5958/0976-1926.2022.00012.2
- Singh A, Verma SK, Prasad G, Kumar A, Sharma PC, Singh AK (2022) Elucidating genetic diversity and population structure in jamun [Syzygium cumini(Skeels)] using morpho-physiological traits and CAAT box-derived polymorphism. South Afri J Bot. 51, 454:464. https://doi.org/10.1016/j.sajb.2022.10.023
- Singh M, Kaur K, Sharma S, Paliwal A, Singh M, Aminedi R, Kaur V and Randhawa G (2022) Assessment of risk of GM contamination in flaxseed accessions imported from Canada: A case study to restrict the unauthorized GM events from entering India. Plant Genetic Resources: Characterization and Utilization, 20 (1), 76-78. doi:10.1017/S1479262122000132
- Singh R, Mahato AK, Singh A, Kumar R, Singh AK, Kumar S, Marla SS, Kumar A and Singh NK (2022) TinoTranscriptDB: A Database of Transcripts and Microsatellite Markers of Tinosporacordifolia, an Important Medicinal Plant. DOI: 10.3390/ genes13081433.
- Singh S, Kaur A, Kakkar P, Dhar S, Madduru D, Yasin JK, Banerjee A, Suravajhala R. (2022) Post-COVID-19 Pandemic Impact Assessment of Bioinformatics and Women Bioinformaticians: A Realm of Possibilities or Gloom-Ridden. The Open COVID Journal,2(1).
- Singh R and R Kumar (2022) Genomic Resource Generation in Medicinal and Aromatic Plants. Indian J. Plant Genet. Resources 35(3): 204–212.
- Verma SK, Mittal S, Gayacharan, Wankhede DP,Parida SK, Chattopadhyay D, Prasad G, Mishra DC, Joshi DC, Singh M, Singh K and Singh AK (2022) Transcriptome Analysis Reveals Key Pathways and Candidate Genes Controlling Seed Development and Size in Ricebean (Vigna umbellata). Frontiers in Genetics. 12:791355. doi: 10.3389/fgene.2021.791355.
- Vikas VK, Pradhan AK, Budhlakoti N, Mishra D, Chandra T, Bhardwaj SC, Kumar S, Sivasamy M, Jayaprakash P, Nisha R, Shajitha P, Peter J, Geetha M, Mir R, Singh K and Kumar S* (2022)Multi-locus genome-wide association studies (ML-GWAS) reveal novel genomic regions associated with seedling and adult plant stage leaf rust resistance in bread wheat (Triticumaestivum). Heredity.DOI: 10.1038/s41437-022-00525-1.
- Wankhede D.P., Singh R, Rajkumar S (2022) Genomic Tools in Plant Genetic Resource Management. Indian Journal of Plant Genetic Resources 35(3):194-199. DOI: 10.5958/0976-1926.2022.00068.7
- Yadav A, Chandra T, and Kumar S (2022) Evaluation of differentially expressed genic marker linked resistance gene analog (RGAs) effective at seedling stage against spot blotch in wheat. Cereal Research Communications.DOI: 10.1007/s42976-022-00285-y.
- Yadav B, Kaur V, Narayan OP, Yadav SK, Kumar A and Wankhede DP (2022) Integrated omics approaches for flax improvement under abiotic and biotic stress: Current status and future prospects. Frontiers in Plant Sciences. 13:931275. https://doi:10.3389/fpls.2022.931275.
- Yadav S, Yadava YK, Kohli D, Meena S, Kalwan G, Bharadwaj C, Gaikwad K, Arora A, Jain PK (2022). Genome-wide identification, in silico characterization and expression analysis of the RNA helicase gene family in chickpea ( arietinum L.). Scientific Reports, 12(1):9778. https://doi.org/10.1038/s41598-022-13823-9
- Yadav S, Yadava YK, Kohli D, Meena S, Paul V, Jain PK (2022). Genome-wide identification and expression analysis of the GRASgene family in response to drought stress in chickpea (Cicer arietinum). 3 Biotech, 12(3):64. DOI: 10.1007/s13205-021-03104-z
- Yasin JK, Mishra BK, Arumugam Pillai M, Chinnusamy V. (2022) Physical map of lncRNAs and lincRNAs linked with stress responsive miRs and genes network of pigeonpea (Cajanus cajan). Journal of Plant Biochemistry and Biotechnology, 31(2), 271-292.
2021
- Andrew-Peter-Leon MT, Selvaraj R, Kumar KK, Muthamilarasan M, Yasin JK, Pillai MA. (2021) Loss of function of OsFBX267 and OsGA20ox2 in rice promotes early maturing and semi-dwarfism in γ-irradiated IWP and genome-edited Pusa Basmati-1. Frontiers in Plant Science, 1968.
- Bansal S, Sharma K, Gautam V, Lone AA, Malhotra EV, Kumar S,Singh R (2021) A Comprehensive Review of Buniumpersicum: AValuable Medicinal Spice. Food Reviews International. DOI:10.1080/87559129.2021.1929305(IF: 6.04)
- Bangar P, N Tyagi, B Tiwari, S Kumar,P Barman R Kumari, , A B Gaikwad K V Bhatand A Chaudhury (2021).Identification and characterization of SNPs in released, landrace and wild accessions of mungbean (Vignaradiata) using whole genome resequencing. Journal of crop Science and Biotechnology 24:163-165. DOI: 1007/s12892-020-00067-0
- Chaurasia S, Singh AK, Kumar A, Songachan LS, Yadav MC, Kumar S, Kumari J, Bansal R, Sharma PC and Singh K (2021) Genome-wide association mapping reveals key genomic regions for physiological and yield-related traits under salinity stress in wheat (Triticum aestivum L.). Genomics 113: 3198-3215. doi: 1016/j.ygeno.2021. 07.014.
- Danakumara T, Kumari J, Singh AK, Sinha SK, Pradhan AK, Sharma S, Jha SK, Bansal R, Kumar S, Jha GK, Yadav MC and Prasad PVV (2021) Genetic dissection of seedling root system architectural traits in a diverse panel of hexaploid wheat through multi-locus genome-wide association mapping for improving drought tolerance. International Journal of Molecular Sciences 22, 7188. doi: 3390/ijms22137188
- Dev R, SK Singh, R Singh, AK Singh, VB Patel, M Alizadeh, K Motha and K Kumar (2021) Assessment of genetic diversity in gamma rays irradiated mutants of four grape genotypes based on RAPD and SSR markers. Indian Journal of Horticulture 78(1): 17-24.
- Gowthami R, Sharma N, Gangopadhyay KK, Rajkumar S, Pathania P and Agrawal A (2021) Cryopreservation of pollen of Abelmoschus moschatus Subsp. Moschatus as an aid to overcome asynchronous flowering for wide hybridization with cultivated Okra [A. Esculentus (l.) Moench] Cryo Letters 42(4): 233-244. https://pubmed.ncbi.nlm.nih.gov/35363843/
- Goyanka J, Yadav MC, Kumari K, Tiwari S and Kumar A (2021). Phenotypic characterization reveals high extent of genetic variation in maize (Zea mays) landraces of North-Eastern and North-Western Himalayan regions of India. Indian Journal of Plant Genetic Resources 34 (3): 389-403. doi: 10.5958/0976-1926.2021.00032.2.
- Khannetah KR, Ramchander S, Leon MA, Shoba D, Saravanan S, Kannan R, Yasin JK, Pillai MA. (2021) Genetic diversity analysis in indigenous rice (Oryza sativa L.) germplasm for bacterial leaf blight (Xanthomonas oryzae pv. oryzae)(BB) using resistance genes-linked markers. Euphytica, 217(7), 145.
- Kumar D, Sharma S, Sharma R, Pundir S, Singh VK, Chaturvedi D, Singh B, Kumar S and Sharma S (2021)Genome wide association study in hexaploid wheat identifies novel genomic regions associated with resistance to root lesion nematode (Pratylenchusthornei).Scientific Reports.DOI: 10.1038/s41598-021-80996-0.
- Kumar R, DasS,Mishra M, Roy Choudhury D, Sharma K, Kumari A, Singh* R (2021)Emerging roles of NAC transcription factor in medicinal plants:progress and prospects. 3 Biotech 11(10):425. (https://doi.org/10.1007/s13205-021-02970-x).(IF: 2.89)
- Lone JK, Lekha MA, Bharadwaj RP, Ali F, Pillai MA, Wani SH, Yasin JK, Chandrashekharaiah KS. (2021) Multimeric Association of Purified Novel Bowman-Birk Inhibitor From the Medicinal Forage Legume Mucuna pruriens (L.) DC. Frontiers in Plant Science. 25;12:772046.
- Lyngkhoi F, N Saini , AB Gaikwad, T Napolean, P Verma , C Silvar , S Yadav and A Khar(2021) Genetic diversity and population structure in onion (Allium cepa) accessions based on morphological and molecular approaches. PhysiolMolBiol Plants 27(11):2517-2532
- Malav PK , A Pandey, V Gupta, Ashok Kumar, KC Bhatt, A Raina, J Akhtar, A B Gaikwad, O S Ahlawat and R Dhaka (2021). The diversity in holy basil(Ocimumtenuiflorum) germpalsm in India. IJAS 90(10):1937-45
- Nikhil HN, AK Goswami, SK Singh, C Kumar, S Goswami,R Singh, C Bharadwaj and NKMaurya (2021) Assessment of morpho-genetic diversity of guava (Psidium guajava)hybrids and genotypes. Indian Journal of Agricultural Sciences. 91(11):1640-45(IF: 0.37)
- Pandey A and Rajkumar S (2021) A new potential variety of cultivated melon (Cucumis melo L.) from north western India. Genet Resour Crop Evol68: 785–794. DOI:1007/s10722-020-00997-2
- Pandey A, Madhav Rai K, Malav PK, Rajkumar S (2021) Allium negianum (Amaryllidaceae): a new species under subg. Rhizirideum from Uttarakhand Himalaya, India. PhytoKeys 183: 77–93. doi: 3897/phytokeys.183.65433
- Patidar A, Yadav MC, Kumari J, Tiwari S, Kushwah MK, Harun M, Paul V and Tomar BS (2021) Morpho-physiological characterization of bread wheat accessions for heat stress tolerance under late sown conditions of North-Western plain zone of India. Indian Journal of Plant Genetic Resources 34: 258-273. doi: 5958/ 0976-1926. 2021.00025.5.
- Phogat BS, Kumar S, Kumari J, Kumar N, Pandey AC, Singh TP; Kumar S, Tyagi RK, Jacob SR, Singh AK, Srinivasan K, Radhamani J, Bisht IS, Archak S, Karale M, Sharma P, Yadav M, Joshi U, Mishra P, Kumari G, Aftab T, Gambhir R, Gangopadhyay KK, YS Rathi, Pal N, Sharma RK, Yadav SK, Bhatt KC, Singh B, Prasad TV, Solanki YPS, Singh D, Dutta M, Yadav MC, Rana JC and Bansal KC (2021) Characterization of wheat germplasm conserved in the Indian National Genebank and establishment of a composite core collection. Crop Science 61(1) 604-620. doi: 1002/csc2.20285.
- Priyadarshini P, Kohli D, Yadav S, Srinivasa N, Bharadwaj C, Anjoy P, Gaikwad K, Jain PK (2021). Quantitative detection of pathogen load of Fusarium oxysporumsp. ciceris infected wilt resistant and susceptible genotypes of chickpea using intergenic spacer region-based marker. Physiol. Mol. Plant Pathol,114, 101622. https://doi.org/10.1016/j.pmpp.2021.101622
- Rao, P.G., Behera, T.K., Gaikwad, A.B. et al.Genetic analysis and QTL mapping of yield and fruit traits in bitter gourd (Momordicacharantia). Sci Rep 11, 4109 (2021). https://doi.org/10.1038/s41598-021-83548-8
- Rathod V, TK Behera, AD Munshi, AB Gaikwad, S Singh, ND Vinay, G Boopalakrishnan and GS Jat (2021). Developing partial interspecific hybrids of Momordicacharantia x Momordicabalsaminaand their advance generations. ScientiaHorticulturae 281 (109985)
- Roy Choudhury D, R Kumar, V Devi S, K Singh, N K Singh, R Singh*(2021)Identification of a diverse core set panel of rice from the east coast region of India using SNP markers. Frontiers in Genetics.12:726152. DOI: 10.3389/fgene.2021.726152(IF: 4.77)
- Sarawgi AK, AK Pachauri, S Vimala Deviand R Singh(2021)Identification of Unique Type of Decorticated Grain Colour in Rice Designated as “Potato Green Colour”. Indian J. Plant Genet. Resour. 34(1): 79–81.(IF: 0.00)
- Singh M, Sodhi KK, Paliwal A, Sharma S and Randhawa G (2021) Efficient DNA extraction procedures for processed food derivatives – A critical step to ensure quality for GMO analysis. Food Analytical Methods, 14: 2249-2261. https://doi.org/10.1007/s12161-021-02051-y
- Sridhar R, M Srivastav, SK Singh, AK Mahto, N Singh, A Nagaraja, R Singh and NK Singh (2021) New genomic markers for marker assisted breeding in mango (Mangiferaindica). The Journal of Horticultural Science & Biotechnology. 96(5): 614-623.
- Srivastav M, SK Singh, J Prakash, R Singh, N Sharma, S Ramchandra, R Devi, A Gupta, AK Mahto, PK. Jayaswal, S Singh and NK Singh (2021) New hyper-variable SSRs for diversity analysis in mango (Mangiferaindica). Indian J. Genet., 81(1):119-126.
- Tripathi K, Parihar AK, Murthy N, Revanasidda, Wankhede DP, Singh N, Deshpande SK, Kumar A (2021) Identification of a unique accession in cowpea with dense pubescence. Journal of Food Legumes 33(4): 278-279.
- Vanniarajan C, Magudeeswari P, Gowthami R, Indhu SM, Ramya KR, Monisha K, Pillai MA, Verma N, Yasin JK. (2021) Assessment of genetic variability and traits association in pigeonpea [Cajanus cajan (L.) Millsp.] germplasm. Legume Research, 46(10), 1280-1287.
- Wani SH, Vijayan R, Choudhary M, Kumar A, Zaid A, Singh V, Kumar P, Yasin JK. (2021) Nitrogen use efficiency (NUE): elucidated mechanisms, mapped genes and gene networks in maize (Zea mays). Physiology and Molecular Biology of Plants, 1-17.
2020
- Marla SS, Mishra P, Maurya R, Singh M, Wankhede DP, Kumar A, Yadav MC, Subbarao N, Singh SK, Kumar R (2020) Refinement of Draft Genome Assemblies of Pigeonpea (Cajanus cajan) Frontiers in Genetics doi.org/10.3389/fgene.2020.607432.
- Rani K, Raghu BR, Jha SK, Agarwal P, Mallick N, M. Niranjana, J. B. Sharma· A. K. Singh N. K. Sharma Rajkumar S. M. S. Tomar Vinod 2020. A novel leaf rust resistance gene introgressed from Aegilopsmarkgrafii maps on chromosome arm 2AS of wheat. Theoretical and Applied Genetics (2020). 133: 2685–2694. DOI: 10.1007/s00122-020-03625-w
- Rohini MR, Sankaran M, Rajkumar S. Prakash K, Gaikwad A, Chaudhury R and Malik SK (2020) Morphological characterization and analysis of genetic diversity and population structure in Citrus × jambhiri Lush. using SSR markers. Genet Resour Crop Evol 67, 1259–1275. DOI:1007/s10722-020-00909-4
- Singh K., Gupta K., Tyagi V., Rajkumar (2020) Plant genetic resources in India: management and utilization. Vavilov Journal of Genetics and Breeding 24(3):306-314. doi: 10.18699/VJ20.622
- Singh M, Pal D, Sood P and Randhawa GJ (2020) Construct-specific loop-mediated isothermal amplification: Rapid detection of genetically modified crops with insect resistance or herbicide tolerance. Journal of AOAC International, 103 (5): 1191-1200. doi: 10.1093/jaoacint/qsaa043
- Singh M, Randhawa GJ, Bhoge RK, Sushmita, Kak A and O Sangwan (2020) Monitoring adventitious presence of transgenes in cotton collections from genebank and experimental plots: Ensuring GM-free conservation and cultivation of genetic resources. Agricultural Research, 9, 469-476. https://doi.org/10.1007/s40003-019-00449-z
- Yadav SK, Santosh Kumar VV, Verma RK, Yadav P, Saroha, Wankhede DP, Chaudhary B, Chinnusamy V (2020) Genome-wide identification and characterization of ABA receptor PYL gene family in rice. BMC Genomics (2020) 21:676. https://doi.org/10.1186/s12864-020-07083-y.
- Yuvraj Y, Lavanya GR, Arya L, Verma M, Rajput R (2020) (Phenotype based selection in kodo millet (Paspalum scrobiculatum) to identify elite accessions. Agricultural Science Digest – A Research Journal 40(4): 357-363. DOI : 10.18805/ag.D-5084
- Yasin JK, Mishra BK, Pillai MA, Verma N, Wani SH, Elansary HO, El-Ansary DO, Pandey PS, Chinnusamy V. (2020) Genome wide in-silico miRNA and target network prediction from stress responsive Horsegram (Macrotyloma uniflorum) accessions. Scientific Reports, 10(1), 17203.
- Singh S, Mahato AK, Jayaswal PK, Singh N, Dheer M, Goel P, Raje RS, Yasin JK, Sreevathsa R, Rai V, Gaikwad K. (2020). A 62K genic-SNP chip array for genetic studies and breeding applications in pigeonpea (Cajanus cajan L. Millsp.). Scientific reports, 10(1), 4960.
- Kannepalli A, Pengani KR, Yasin JK, Paul S, Tyagi S, Venkadasamy G, Sharma M, Karivaradharajan S. (2020). Genome Assembly of Azotobacter chroococcum Strain W5, a Free-Living Diazotroph Isolated from India. Microbiology Resource Announcements, 9(20), 10-1128. https://doi.org/10.1128/mra.00259-20
- Kumar S, Kumari J, Bhusal N, Pradhan AK, Budhlakoti N, Mishra DC, Chauhan D, Kumar S, Singh AK, Reynolds MP, Singh GP, Singh K and Sareen S (2020) Genome-wide association study reveals genomic regions associated with ten agronomical traits in wheat under late-sown conditions.Frontiers in Plant Science.DOI: 10.3389/fpls.2020.549743.
- Pradhan AK, Kumar S, Singh AK, Budhlakoti N, Mishra DC, Chauhan D, Mittal S, Grover M, Kumar S, Gangwar OP, Kumar S, Gupta A, Bhardwaj SC, Rai A and Singh K (2020) Identification of QTLs/DefenseGenes Effective at Seedling Stage Against Prevailing Races of Wheat Stripe Rust in India. Frontiers in Genetics.DOI: 10.3389/fgene.2020.572975.
- Vikas VK, Kumar S, Archak S, Tyagi RK, Kumar J and Bansal KC (2020) Screening of 19,460 genotypes of wheat species for resistance to powdery mildew and identification of potential candidates using focused identification of germplasm strategy (FIGS). Crop Science.DOI: 10.1002/csc2.20196.
- Singh AK, Singh N, Kumar S, Kumari J, Singh R, Gaba S, Yadav MC, Grover M, Chaurasia S and Kumar R (2020) Identification and evolutionary analysis of polycistronic miRNA clusters in domesticated and wild wheat.DOI: 10.1016/j.ygeno.2020.01.005.
- Singh S, Mishra VK, Kharwar RN, Budhlakoti N, Ahirwar RN, Mishra DC, Kumar S, Chand R, Kumar U, Kumar S and Joshi AK (2020) Genetic characterization for lesion mimic and other traits in relation to spot blotch resistance in spring wheat. PLoS ONE.DOI:10.1371/journal.pone.0240029.
- Phogat BS, Kumar S, Kumari J, Kumar N, Pandey AC, Singh TP, Kumar S and Bansal KC (2020) Characterization of wheat germplasm conserved in the Indian National Genebank and establishment of a composite core collection. Crop Science.DOI: 10.1002/csc2.20285.
- UpadhyayD, Budhlakoti N, Singh AK, Bansal R, Kumari J, Chaudhary N, Padaria JC, Sareen S and Kumar S (2020)Drought tolerance in Triticum aestivum Genotypes associated with enhanced antioxidative protection and declined lipid peroxidation.3 Biotech.DOI:10.1007/s13205-020-02264-8.
- Shipra Deo, Anto James, Sanjeev Kumar Singh, CB Singh and Mukesh Kumar Rana (2020). Molecular diversity analysis and cultivar identification using simple sequence repeat (SSR) markers in soybean [Glycine max (L.) Merrill] International Journal of Current Microbiology and Applied Sciences 9(4).
- Shipra Deo, Anto James, Sanjeev Kumar Singh, C.B. Singh and Mukesh Kumar Rana. 2020.Development of Multiplex Microsatellite Marker Sets in Soybean [Glycine max (L.) Merr.].International Journal of Current Microbiology and Applied Sciences 9(10): doi: https://doi.org/10.20546/ijcmas.2020.910.xx
- Marla S, Mishra P, Maurya R, Singh M, Wankhede DP, Kumar A, Yadav MC, Rao NS, Singh SK and Kumar R (2020) Refinement of draft genome assemblies of pigeonpea (Cajanus cajan). Frontiers in Genetics 11:1-12. https://doi.org/3389/fgene.2020.607432.
- Kumar N, Bharadwaj C, Soni A, Sachdeva S, Yadav MC, Pal M, Soren KR, Meena MC, Roorkiwal M, Varshney RK and Rana M (2020). Physio-morphological and molecular analysis for salt tolerance in chickpea (Cicer arietinum). Indian Journal of Agricultural Sciences 90(4): 804-808.
- Tiwari S, Yadav MC, Dikshit N, Yadav VK, Pani DR and Lata M (2020). Morphological characterization and genetic identity of crop wild relatives of rice (Oryza sativa L.) collected from different ecological niches of India. Genetic Resources and Crop Evolution 67: 2037–2055. https://doi.org/ 10.1007/s10722-020-00958-9.
- Singh AK, Singh N, Kumar S, Kumari J, Singh R, Gaba S, Yadav MC, Grover M, Chaurasia S and Kumar R (2020). Identification and evolutionary analysis of polycistronic miRNA clusters in domesticated and wild wheat. Genomics 112: 2334-2348. https://doi.org/10.1016/j.ygeno.2020.01.005
- Kumar R, C Kuma, R Paliwal, DR Choudhury, I Singh, A Kumar, A Kumari and R Singh (2020) Development of Novel Genomic Simple Sequence Repeat (g-SSR) Markers and Their Validation for Genetic Diversity Analyses in Kalmegh[ Andrographis Paniculata (Burm. F.) Nees]. Plants 9(12):1734.
- Kumar C, R Kumar, SK Singh, AK Goswami, A.Nagaraja, R Paliwal, R Singh (2020). Development of novel g-SSR markers in guava (Psidium guajava) cv. Allahabad Safeda and their application in genetic diversity, population structure and cross species transferability studies.PLoS ONE 15(8): e0237538.
- Singh A, YSingh, A Mahato, P Jayaswal, S Singh, RSingh, N Yadav, A Singh, P Singh, R Singh, R Kumar, E Septiningsih, H Balyan, N K Singh (2020). Allelic sequence variation in the Sub1A, Sub1B and Sub1C genes among diverse rice cultivars and its association with submergence tolerance. Scientific Reports 10:8621 (https://doi.org/10.1038/s41598-020-65588-8).
- Singh AK, N Singh, S Kumar, J Kumari, R Singh,S Gaba, MC Yadav, MGrover, S Chaurasia, RKumar (2020)Identification and evolutionary analysis of polycistronic miRNA clusters indomesticated and wild wheat. Genomics 112: 2334-2348.(IF: 4.31)
2019
- Arya L, Verma M, Singh SK, Verma RPS (2019) Spatio-temporal genetic diversity in Indian barley (Hordeum vulgare L.) varieties based on SSR markers. Indian J Exp Biol 57:545-552
- Das R, V Arora, S Jaiswal, MA Iquebal, UB Angadi, S Fatma, R Singh, S Shil, A Rai and D Kumar (2019). PolyMorphPredict: A universal web tool for rapid polymorphic microsatellite marker discovery for whole genome and transcriptome data. Frontiers in Plant Sciences. 9: 1-10. doi: 10.3389/fpls.2018.01966. (IF: 6.63)
- Gayacharan, Bisht IS, Bhardwaj R, Rana JC, Singh AK, and Yadav MC (2019). Nutritional diversity of elite rice landraces from subsistence-oriented farming systems. Indian Journal of Plant Genetic Resources 32(1): 18-27. https://doi.org/5958/0976-1926.2019.00003.2
- Kaur V, Yadav SK, Wankhede DP, Pulivendula P, Kumar A, Chinnusamy V (2019) Cloning and characterization of a gene encoding MIZ1, a domain of unknown function protein and its role in salt and drought stress in rice. Protoplasma. doi: 10.1007/s00709-019-01452-5.
- Kiran Babu P, J Radhamani, S Rajkumar and RK Tyagi. (2019). Note on Diversity in Legumes and Oilseeds and their Wild Relatives in Eastern Ghats of India: Utilization and Conservation Concerns. Indian J. Plant Genet. Resour. 32(1): 43-53. DOI:5958/0976-1926.2019.00006.8
- Kuldeep S, Kumar S, Rajkumar S, Singh Mohar, Gupta K (2019) Plant genetic resources management and pre-breeding in genomics era. Indian J. Genet., 79(1) Suppl. 117-130. DOI: 10.31742/IJGPB.79S.1.1
- Kumar C, SK Singh, R Singh, KK Pramanick, MK Verma, M Srivastav, G Tiwari, DR Choudhury (2019) Genetic diversity and population structure analysis of wild Malusgenotypes including the crabapples ( baccata (L.) Borkh. & M. sikkimensis (Wenzig) Koehne ex C. Schneider) collected from the Indian Himalayan region using microsatellite markers. Genetic Resources and Crop Evolution 66(6):1311-1326.
- Kumar C, SK Singh, R Singh,MK Verma, KK Pramanick, M Srivastav, R Kumar, JK Verma and NK Negi (2019). Analysis of genetic diversity and population structure of the Indigenous and exotic wild Malus species using ISSR markers. Indian Journal of Agricultural Sciences 89(7):1096-1102.
- Kumar S, BS Phogat, Vikas VK, AK Sharma, MS Saharan, AK Singh, J Kumari, R Singh, SR Jacob, GP Singh, M Sivasamy, P Jayaprakash, Madhu Meeta, JP Jaiswal, Deep Shikha, BK Honrao, IK Kalappanavar, PC Mishra, SP Singh, SS Vaish and VA Solanki (2019). Mining of Indian wheat germplasm collection for adult plant resistance to leaf rust. PLoS ONE 14(3):e0213468. DOI:10.1371/journal.pone.0213468
- Kumar U, Kumar S, Prasad R, Röder MS, Kumar S, Chand R, Mishra VKand Joshi AK (2019) Genetic Gain on Resistance to Spot Blotch of Wheat by Developing Lines with Near Immunity Crop Breeding, Genetics & Genomics, 1:e190017.DOI: 20900/cbgg20190017.
- Kumari R, DP Wankhade, A Bajpai, A Maurya, K Prasad, D Gautam, P Rangan m Latha, JK John, A Suma, KV Bhat and AB Gaikwad (2019).Genome wide identification and characterization of microsatellite markers in black pepper (Piper nigrum): A valuable resource for boosting genomics applications PLOS ONE 14(12): e0226002. https://doi.org/10.1371/journal.pone.0226002
- Lyngkhoi F, A Khar, M Mangal, AB Gaikwad, T Napolean (2019). Expression analysis and association of bulbing to FLOWERING LOCUS T(FT) gene in short day onion (Allium cepa). Indian J of Genet. 79(1)77-81
- Mahfooz S, Srivastava A, Yadav MC and Tahoor Z (2019). Comparative genomics in phytopathogenic prokaryotes reveals the higher relative abundance and density of long-SSRs in the smallest prokaryotic genome. 3 Biotech. 9:340. https://doi.org/10.1007/s13205-019-1872-8.
- Pandey A, K Pradheep, AB Gaikwad, R Gupta, PK Malav and M Rai (2019).Systematics study on a morphotype of Allium tuberosumRottler ex Spreng. (Alliaceae) from Ladakh , India. Indian J. Plant Genet. Resour. 32(2):223-231
- Raina SK, Raskar N, Aher L, Singh AK, Wankhede DP, Rane J and Minhas PS (2019) Variations in ethylene sensitivity among mungbean [Vigna radiata (L.) Wilczek] genotypes exposed to drought and waterlogging stresses. Turk J Bot. 43: doi:10.3906/bot-1902-33
- Sareen S, Bhusal N, Kumar M, Bhati PK, Munjal R, Kumari J, Kumar S, Sarial AK (2019) Molecular genetic diversity analysis for heat tolerance of indigenous and exotic wheat genotypes Plant Biochem. Biotechnol. DOI: 10.1007/s13562-019-00501-7.
- Singh M, Bhoge RK, Nain S and Randhawa GJ (2019) Loop-mediated isothermal amplification: A rapid detection method for rice actin and nopaline synthase promoters in genetically modified crops. Journal of Plant Biochemistry & Biotechnology, 28 (3): 353-356. https://doi.org/10.1007/s13562-018-0479-1
- Singh M, Pal D, Sood P and Randhawa GJ (2019) Loop-mediated isothermal amplification assays: Rapid and efficient diagnostics for genetically modified crops. Food Control, 106: 106759. https://doi.org/10.1016/j.foodcont.2019.106759
2018
- Ahirwar RN, Mishra VK, Chand R, Budhlakoti N, Mishra DC, Kumar S, et al.(2018)Genome-wide association mapping of spot blotch resistance in wheat association mapping initiative (WAMI) panel of spring wheat (Triticumaestivum).PLoS ONE.DOI: 10.1371/journal.pone.0208196.
- Bangar P , A Chaudhury , S Umdale, R Kumari, B Tiwari, S Kumar, A B Gaikwad and K V Bhat (2018) Detection and characterization of polymorphic simple sequence repeats markers for the analysis of genetic diversity in Indian mungbean [Vigna radiata (L.) Wilczek]. Indian J. Genet., 78(1): 111-117 (2018) DOI: 10.5958/0975-6906.2018.00013.5
- Gaikwad AB, S Saxenaand S Archak (2018). Development of AFLP markers and molecular diversity analysis in saffron (Crocus sativus L.). ActaHortic. 1200, 1-6 https://doi.org/10.17660/ActaHortic.2018.1200.1
- Gayacharan, Bisht IS, Pandey A, Yadav MC, Singh AK, Pandravada SR and Rana JC (2018). Population structure of some indigenous aromatic rice (Oryza sativa) landraces of India. Indian Journal of Biotechnology 17 (1): 110-117.
- Kaur V, Kumar S., Yadav R., Wankhede D.P., Aravind J., Radhamani J.,. Rana J.C and Kumar A. (2018) Analysis of genetic diversity in Indian and exotic linseed germplasm and identification of trait specific superior accessions. Journal of Environmental Biology 39-5:702-709. http://doi.org/10.22438/jeb/39/5/MRN-849.
- Kuldeep K, Gore PG, Gayacharan, Kumar S, Bhalla S and Bisht IS (2018) Population structure and genetic diversity of wheat landraces from North-Western Indian Himalya.Indian J. Plant Genet. DOI: 10.5958/0976-1926.2018.00020.7.
- Kumar C, Singh SK, Singh R, Pramanick KK,Verma MK and Srivastava M (2018) Genetic diversity and population structure studies of the wild apple (Malus) genotypes using RAPD markers. Indian Journal of Horticulture. 75(4):557-564.
- Kumar C, SK Singh, KK Pramanick, MK Verma, M Srivastav, R Singh, C Bharadwaj, KC Naga (2018)Morphological and biochemical diversity among the Malus species including indigenous Himalayan wild apples. Scientia Horticulturae 233: 204–219
- Kumar S, Kumari J, Bansal R, Kuri BR, Upadhyay D, Srivastava A, Rana B, Yadav MK, Sengar RS, Singh AK and Singh R (2018) Multi-environmental evaluation of wheat genotypes for drought tolerance. Indian J. Genet. Plant Breed.DOI : 10.5958/0975-6906.2018.00004.4.
- Kumar S, J Kumari, R Bansal, BR Kuri, D Upadhyay, P K Bhati, A Srivastava, B Rana, MK Yadav, RS Sengar, AK Singhand R Singh (2018) Identification of stable drought tolerant wheat genotypes under multi-environmental trials. Indian Journal of Genetics and Plant Breeding,78(1): 26-35.
- Kumar S, S Vats, P Barman, N Tyagi, R Kumari, P Bangar, B Tiwari, S Sachdeva, A B Gaikwadand K V Bhat (2018). Whole genome SNP identification and validation in Cucumis melo L. cultivars using genome resequencing approach. Indian J Genet. and Pl. Br., 78(4): 478-486
- Kumari J, Kumar S, Singh N, Vaish SS, S Das, Gupta A and Rana JC (2018) Identification of New Donors for Spot Blotch Resistance in Cultivated Wheat Germplasm. Cereal Research Communications.DOI: 10.1556/0806.46.2018.028.
- Kumari S, N Arumugam, R Singh, M Srivastav, S Banoth, AC Mithra, MB Arun, AK Goswami and YJ Khan (2018) Diversity analysis of guava (Psidium guajava) germplasm collection. Indian Journal of Agricultural Sciences 88(3):489-497.(IF: 0.37)
- Mohan H, Arya L, Verma M, Bisht IS, Saha D, Dhillon BS (2018) Morpho-molecular variation in Indian finger millet (Eleusine coracana (L.) Gaertn.) varieties and landraces. Indian J Plant Genet Resour 31(3): 276–285. DOI 10.5958/0976-1926.2018.00032.3
- Motha K, SK Singh, AK Singh, R Singh, M Srivastav, MK Verma and C Bhardwaj (2018)Molecular Characterization and Genetic Relationshipsof Some Stress Tolerant Grape Rootstock Genotypes asRevealed by ISSR and SSR Markers. Plant Tissue Culture& Biotechnology28(1): 77-90.(IF: 0.00)
- Rao PG, TK Behera, AB Gaikwad, AD Munshi, JS Jat and G Boopalakrishnan (2018). Mapping and QTL analysis of gynoecy and earliness in bitter gourd (Momordicacharantia L) using genotyping by sequencing technology. Frontiers in Plant Science doi:10.3389/fpls.2018.01555
- Shaili K, A Nagaraja, Rakesh Singh, Srivastav M, Banoth S, Mithra AC, Arun MB, Goswami AK, Yasin JK. (2018). Diversity analysis of guava (Psidium guajava) germplasm collection. Indian J. Agr. Sciences,88(3), 489-497.
- S Yadav, C Ram, S Singh and MK Rana (2018) Plant Genomic DNA Isolation – the Past and the Present, a Review, Indian J. Plant Genet. Resour. 31(3):315–327.
- Singh AK#, Chaurasia S #, Kumar S #, Singh R, Kumari J, Yadav MC, Singh N, Gaba S and Jacob SR (2018)Identification, analysis and development of salt-responsive candidate gene-based SSR markers in wheat. BMC Plant Biology.DOI: 10.1186/s12870-018-1476-1.
- Singh B, AB Gaikwad, C Ram, S Archak (2018). DNA profiling of pomegranate (Punicagranatum L. ) field genebank semi-feral collection by using ISSR marker. Indian Journal of Plant Genetic Resources, 31(2): 191-193.
- Singh M, Bhoge RK and GJ Randhawa (2018) Loop-mediated isothermal amplification for detection of endogenous Sad1 gene in cotton: An internal control for rapid on-site GMO testing. Journal of AOAC International, 101(5):1657-1660. https://doi.org/10.5740/jaoacint.18-0016
- Thakur AK, Singh KH, Singh L, Nanjundan J, Yasin JK, Singh D. (2018). Patterns of subspecies genetic diversity among oilseed Brassica rapa as revealed by agro-morphological traits and SSR markers. Journal of Plant Biochemistry and Biotechnology, 26, 282-292.
- Tiwari B, S Kalim, N Tyagi, R Kumari, P Bangar, P Barman, S Kumar, A B Gaikwad and K V Bhat (2018). Identification of genes associated with stress tolerance in moth bean [Vignaaconitifolia (Jacq.) Marechal] a stress hardy crop. Physiol. Mol. Biol. Plants doi 10.1007/s 12290-018-0525-4
- Tiwari B, S Kalim, P Bangar, R Kumari, S Kumar, A B Gaikwad and K V Bhat (2018). Physiological, biochemical and molecular responses of thermotolerance in moth bean [Vignaaconitifolia (Jacq.) Marechal]. Turk.J Agric. Forestry. doi 10.3906/tar-1709-1
- Trivedi AK, Arya L, Verma SK, Tyagi RK, Hemantaranjan A, Verma M, Singh VP, Saha D (2018) Molecular profiling of foxtail millet (Setaria italica (L.) P. Beauv) from Central Himalayan region for genetic variability and nutritional quality. J Agricul Sci 156(3):333-341
- Wankhede DP, Aravind J, & Mishra SP (2019) Identification of Genic SNPs from ESTs and Effect of Non-synonymous SNP on Proteins in Pigeonpea. Proc. Natl. Acad. Sci., India, Sect. B Biol. Sci. 89, 2: 595–603. https://doi.org/10.1007/s40011-018-0973-1.
Projects
| S. No. | Project Title | Funding Agency | Principle Investigator | Date of Start | Date of Termination | Budget (Lakhs) | Project Code |
| 1. | NNP (Agri-Genomic Repository and Intelligent Analytical System) of ICAR- Indian Agricultural Research Institute, New Delhi | DBT | Rakesh Singh | 23rd January 2024 | 22nd January 2029 | 43.44 | 1015233 |
| 2. | Enhancing Climate Resilience and Ensuring Food Security with Genome Editing Tool | ICAR | Rakesh Singh | 20th March 2024 | 31st March 2026 | 226.46 | 1015170 |
| 3. | Rationalisation of rice collections originating from major areas of diversity and allele mining in selected unique set of accessions for biotic, abiotic and quality traits using molecular markers. | ICAR | Rakesh Singh | April 2021 | March 2026 | – | 1001381 |
| 4. | Identification and validation of climate-smart germplasm and pre-breeding for abiotic stress tolerance in wheat and rice using crop wild relatives and related species | ICAR | Mahesh Chandra Yadav | April 2021 | March 2026 | 534.00 | OXX04063-ICAR-MY- |
| 5. | Genomics of black pepper, cardamom and fenugreek for relating genes to traits of economic importance | ICAR | Ambika Gaikwad | October 2017 | March 2026 | 139.75 | 1611316024 |
| 6. | Identification of genes/genomic regions associated with Fusarium head blight resistance in wheat | ICAR | Sundeep Kumar | April 2021 | March 2026 | 85.00 | 1004936 |
| 7. | Towards understanding the C3-C4 intermediate pathway in Poaceae and functionality of C4 genes in rice | ICAR | Parimalan R | April 2021 | March 2026 | 150.00 | 1006976 |
| 8. | A detailed foodomics study for food authentication and exploration of nutraceutical potential | NASF | Manjusha Verma | July 2022 | June2025 | 71.063 | 1013764 |
| 9. | Mainstreaming rice landraces diversity in varietal development through genome- wide association studies: A model for large-scale utilization of gene bank collections of rice | DBT | Rakesh Singh | March 2020 | March 2025 | 2227.90 | 1012179 |
| 10. | Development of amaranth core collection using SSR and SNP markers and evaluation of core set for nutritional, yield traits and abiotic stress tolerance | DBT | Rakesh Singh | March 2019 | September 2024 | 212.54 | 1011049 |
| 11. | Development of superior haplotype based near isogenic lines (Haplo-NILs) for enhanced genetic gain in rice | DBT | Rakesh Singh | March 2020 | March 2024 | 157.12 | 1012181 |
| 12. | A National Mission Mode Program on Nutritional improvement of digestible protein content and quality in rice. | DBT | Rakesh Singh | January 2022 | January 2027 | 101.22 | 1013419 |
| 13. | Germplasm characterization and trait discovery in wheat using genomics approaches and its integration for improving climate resilience, productivity and nutritional | DBT | Sundeep Kumar | February 2020 | February 2025 | 127.12 | 1012159 |
| 14. | Germplasm genomics for trait discovery in wheat | DBT | Amit Kumar Singh & Sundeep Kumar | February 2020 | February 2025 | 862.00 | 1012152 |
| 15. | Capacity building in the area of genomic selection and high throughput phenotyping and its integration in national breeding programs | DBT | Sundeep Kumar | February 2020 | February 2025 | 48.00 | 1012158 |
| 16. | Evaluation of wheat germplasm for quality traits | DBT | Sundeep Kumar | February 2020 | February 2025 | 51.36 | 1012156 |
| 17. | Exploiting genetic diversity for improvement of safflower through genomics-assisted discovery of QTLs/Genes associated with agronomic traits | DBT | S Rajkumar | February 2020 | February 2025 | 597.88 | 1012259 |
| 18. | Genotyping of sesame germplasm following high throughput genomics | DBT | Parimalan R | February 2020 | February 2025 | 1000.00 | 16113200037 |
| 19. | National Programme for Quarantine and GM Diagnostics of Genetically Engineered Plant Material (Component-2) | DBT | Monika Singh | September 2020 | September 2025 | 278.56 | 1012550 |
| 20. | Development of reference genome and core set using molecular markers: under network project ‘Leveraging genetic resources for accelerated genetic improvement of linseed using comprehensive genomics and phenotyping approaches’ | DBT | DP Wankhede | February 2020 | February 2025 | 305.95 | 1012140 |
Staff
Scientist

Head of Division
Division of Genomic Resources
Phone: 011-25802884
Email: rakesh.singh2-icar@nic.in, singhnbpgr@yahoo.com

Principal Scientist
Division of Genomic Resources
Phone: 011-25802762
Email: Mahesh.Yadav1-icar@nic.in, mcyadav@yahoo.com, maheshgenome@gmail.com

Principal Scientist
Division of Genomic Resources
Phone:011-25802809
Email: Mukesh.Rana-icar@nic.in

Principal Scientist
Division of Genomic Resources
Phone: 011-25802796
Email: Ambika.Gaikwad-icar@nic.in

Principal Scientist
Division of Genomic Resources
phone:011-25802797
Email: Lalit.Arya-icar@nic.in

Principal Scientist
Division of Genomic Resources
phone:011-25802799
Email: Manjusha.Verma-icar@nic.in, manjusha_v@yahoo.com

Principal Scientist
Division of Genomic Resources
Phone: 011-25802878
Email: Sundeep.Kumar-icar@nic.in

Principal Scientist
Division of Genomic Resources
phone:011-25802805
Email: Rajesh.Kumar13-icar@nic.in, kraj.pgr@gmail.com

Principal Scientist
Division of Genomic Resources
Phone: 011-25802821
Email: Sangita.Bansal-icar@nic.in, sangitabansal@yahoo.com

Principal Scientist
Division of Genomic Resources
phone:011-25802802
Email: s.rajkumar-icar@nic.in, rajnbpgr@gmail.com

Principal Scientist
Division of Genomic Resources
phone:011-25802810
Email: vishnu.kumar-icar@nic.in

Principal Scientist
Division of Genomic Resources
phone:011-25802889
Email: r.parimalan-icar@nic.in, parimalan.r@gmail.com

Principal Scientist
Division of Genomic Resources
phone:011-25802790
Email: amit.singh5@icar.org.in, amitsinghbiotech@gmail.com

Senior Scientist
Division of Genomic Resources
Phone: 011-25802831
Email: susheel.raina-icar@nic.in, Susheelkr76@gmail.com

Senior Scientist
Division of Genomic Resources
phone:011-25802890
Email: yasin.jeshima@icar.org.in

Senior Scientist
Division of Genomic Resources
Phone: 011-25802839
Email: monika.singh-icar@nic.in, monikasinghnbpgr@gmail.com

Senior Scientist
Division of Genomic Resources
Phone: 011-25802875
Email: d.wankhede-icar@nic.in, wdhammaprakash@gmail.com

Senior Scientist
Division of Genomic Resources
phone:011-25802877
Email: M.Priyadarshi-icar@nic.in, madhu74_nbpgr@yahoo.com

Senior Scientist
Division of Genomic Resources
phone:011-25802891
Email: gayacharan-icar@nic.in

Senior Scientist
Division of Genomic Resources
phone:011-25802798
Email: laxmi.sharma-icar@nic.in, laxmi1408@gmail.com

Senior Scientist
Division of Genomic Resources
Phone: 011-25802798
Email: Sheel.Yadav-icar@nic.in, sheel.y85@gmail.com

Scientist
Division of Genomic Resources
phone:011-25802807
Email: shruti.sinha-icar@nic.in
Technical

Chief Technical Officer
Division of Genomic Resources
Phone: 011-25802761
Email: Dikshant.Gautam-icar@nic.in, dikshant7874@hotmail.com, dikshant7874@rediffmail.com

Technical Officer
Division of Genomic Resources
phone:011-25802761
Email: manish.tomar-icar@nic.in, manishtomar028@gmail.com

Senior Technical Assistant
Division of Genomic Resources
phone:011-25802804
Email: Kushaldeep.KaurSodhi-icar@nic.in, kushaldeepkaursodhi@gmail.com



